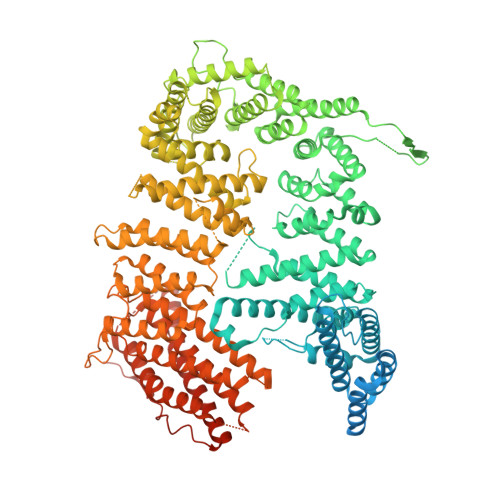
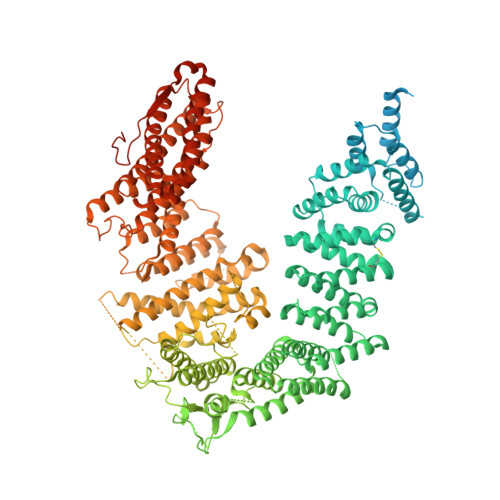
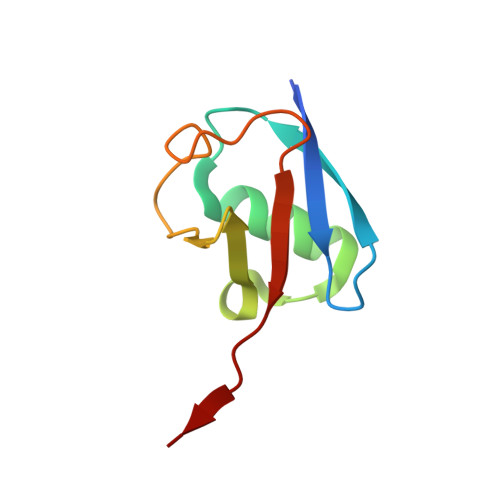
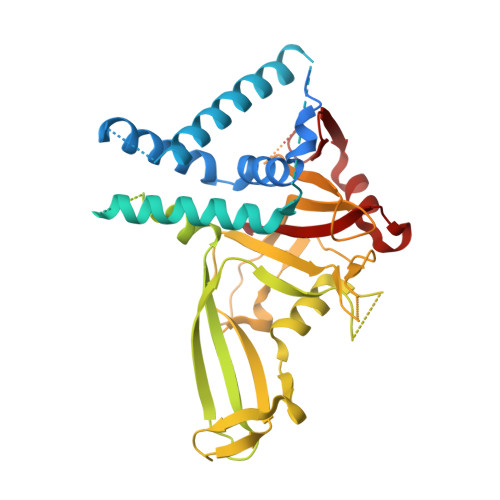
Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Rennie, M.L., Arkinson, C., Chaugule, V.K., Walden, H.(2022) Sci Adv 8: eabq6353-eabq6353
- PubMed: 36170365
- DOI: https://doi.org/10.1126/sciadv.abq6353
- Primary Citation of Related Structures:
7ZH3, 7ZH4, 8A9J, 8A9K - PubMed Abstract:
Repair of DNA damage is critical to genomic integrity and frequently disrupted in cancers. Ubiquitin-specific protease 1 (USP1), a nucleus-localized deubiquitinase, lies at the interface of multiple DNA repair pathways and is a promising drug target for certain cancers. Although multiple inhibitors of this enzyme, including one in phase 1 clinical trials, have been established, their binding mode is unknown. Here, we use cryo-electron microscopy to study an assembled enzyme-substrate-inhibitor complex of USP1 and the well-established inhibitor, ML323. Achieving 2.5-Å resolution, with and without ML323, we find an unusual binding mode in which the inhibitor disrupts part of the hydrophobic core of USP1. The consequent conformational changes in the secondary structure lead to subtle rearrangements in the active site that underlie the mechanism of inhibition. These structures provide a platform for structure-based drug design targeting USP1.
- Institute of Molecular Cell and Systems Biology, College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow, UK.
Organizational Affiliation: