Structural basis for DARC binding in reticulocyte invasion by Plasmodium vivax.
Moskovitz, R., Pholcharee, T., DonVito, S.M., Guloglu, B., Lowe, E., Mohring, F., Moon, R.W., Higgins, M.K.(2023) Nat Commun 14: 3637-3637
- PubMed: 37336887
- DOI: https://doi.org/10.1038/s41467-023-39357-w
- Primary Citation of Related Structures:
8A44 - PubMed Abstract:
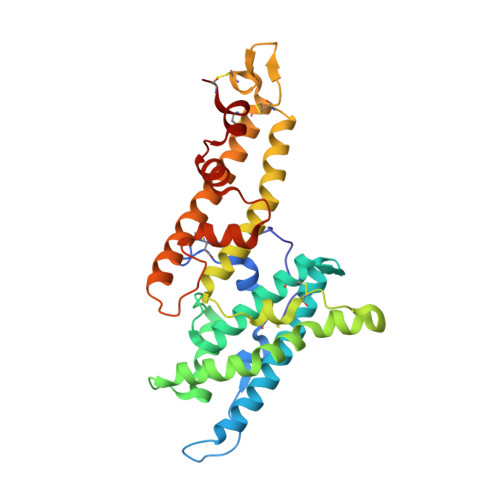
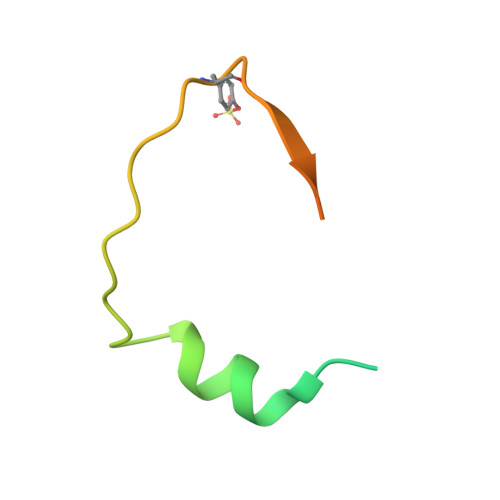
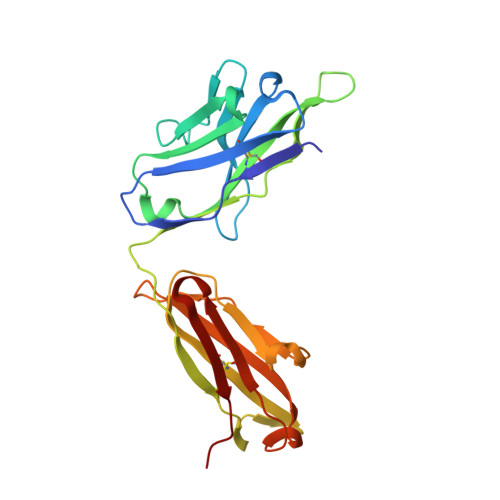
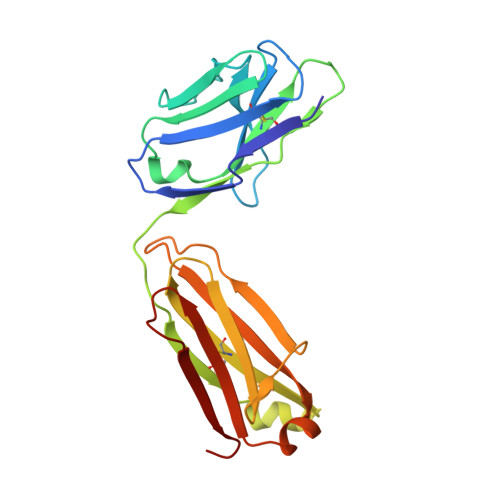
The symptoms of malaria occur during the blood stage of infection, when the parasite replicates within human red blood cells. The human malaria parasite, Plasmodium vivax, selectively invades reticulocytes in a process which requires an interaction between the ectodomain of the human DARC receptor and the Plasmodium vivax Duffy-binding protein, PvDBP. Previous studies have revealed that a small helical peptide from DARC binds to region II of PvDBP (PvDBP-RII). However, it is also known that sulphation of tyrosine residues on DARC affects its binding to PvDBP and these residues were not observed in previous structures. We therefore present the structure of PvDBP-RII bound to sulphated DARC peptide, showing that a sulphate on tyrosine 41 binds to a charged pocket on PvDBP-RII. We use molecular dynamics simulations, affinity measurements and growth-inhibition experiments in parasites to confirm the importance of this interaction. We also reveal the epitope for vaccine-elicited growth-inhibitory antibody DB1. This provides a complete understanding of the binding of PvDBP-RII to DARC and will guide the design of vaccines and therapeutics to target this essential interaction.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU, UK.
Organizational Affiliation: