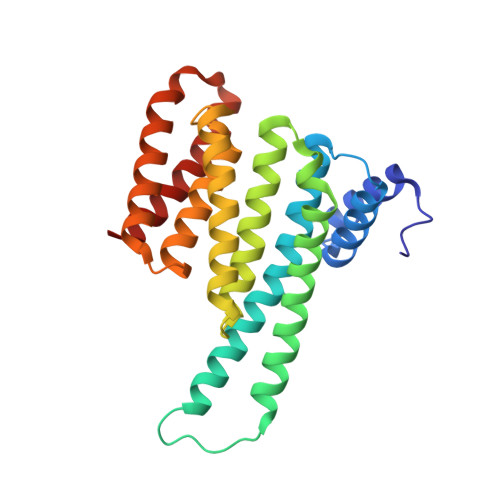
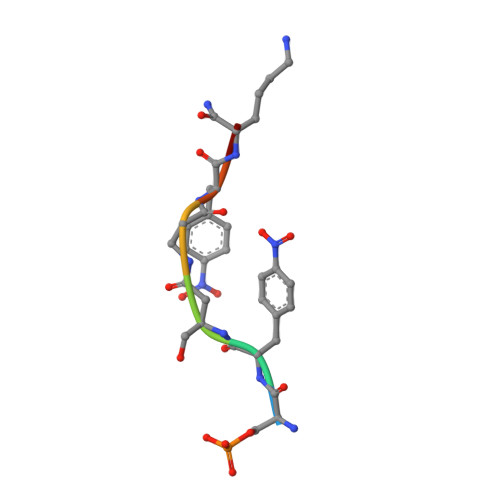
Functional mapping of the 14-3-3 hub protein as a guide to design 14-3-3 molecular glues.
Somsen, B.A., Craenmehr, F.W.B., Liu, W.W., Koops, A.A., Pennings, M.A.M., Visser, E.J., Ottmann, C., Cossar, P.J., Brunsveld, L.(2022) Chem Sci 13: 13122-13131
- PubMed: 36425501
- DOI: https://doi.org/10.1039/d2sc04662h
- Primary Citation of Related Structures:
7ZMU, 7ZMW - PubMed Abstract:
Molecular glues represent an evolution in drug discovery, however, targeted stabilization of protein complexes remains challenging, owing to a paucity of drug design rules. The functional mapping of hotspots has been critical to protein-protein interaction (PPI) inhibitor research, however, the orthogonal approach to stabilize PPIs has not exploited this information. Utilizing the hub protein 14-3-3 as a case study we demonstrate that functional mapping of hotspots provides a triage map for 14-3-3 molecular glue development. Truncation and mutation studies allowed deconvoluting the energetic contributions of sidechain and backbone interactions of a 14-3-3-binding non-natural peptide. Three central 14-3-3 hotspots were identified and their thermodynamic characteristics profiled. In addition to the phospho-binding pocket; (i) Asn226, (ii) Lys122 and (iii) the hydrophobic patch formed by Leu218, Ile219 and Leu222 were critical for protein complex formation. Exploiting this hotspot information allowed a peptide-based molecular glue that elicits high cooperativity ( α = 36) and selectively stabilizes the 14-3-3/ChREBP PPI to be uniquely developed.
- Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology P.O. Box 513 Eindhoven 5600 MB The Netherlands p.cossar@tue.nl l.brunsveld@tue.nl.
Organizational Affiliation: