Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Michalczyk, E., Hommernick, K., Behroz, I., Kulike, M., Pakosz-Stepien, Z., Mazurek, L., Seidel, M., Kunert, M., Santos, K., von Moeller, H., Loll, B., Weston, J.B., Mainz, A., Heddle, J.G., Sussmuth, R.D., Ghilarov, D.(2023) Nat Catal 6: 52-67
- PubMed: 36741192
- DOI: https://doi.org/10.1038/s41929-022-00904-1
- Primary Citation of Related Structures:
7Z9C, 7Z9G, 7Z9K, 7Z9M - PubMed Abstract:
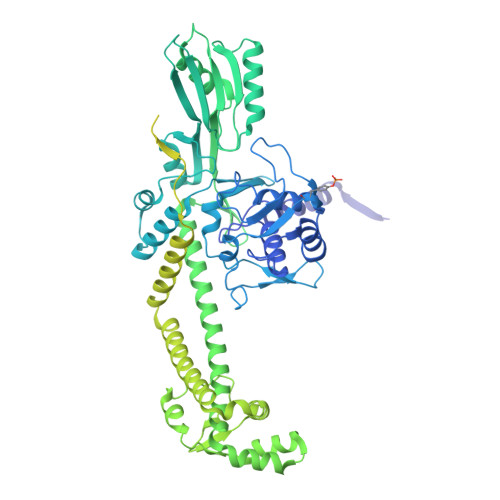
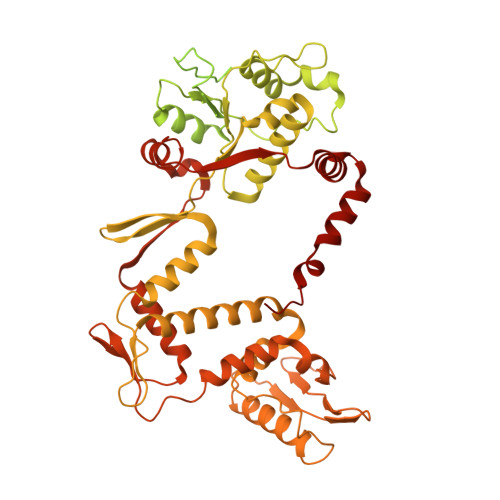
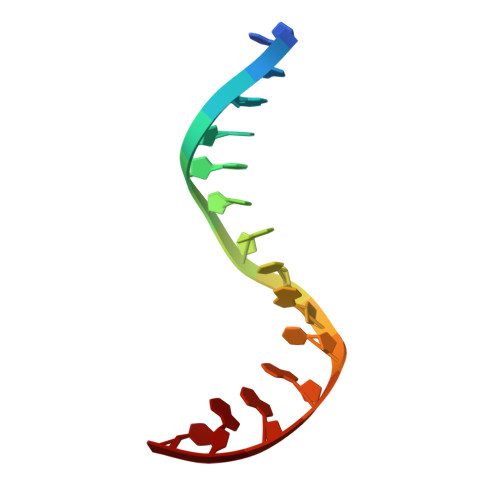
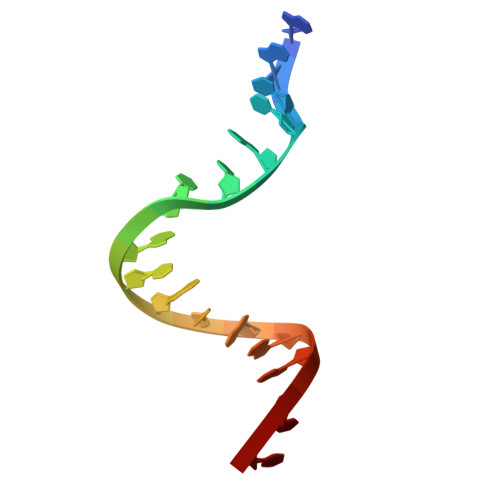
The peptide antibiotic albicidin is a DNA topoisomerase inhibitor with low-nanomolar bactericidal activity towards fluoroquinolone-resistant Gram-negative pathogens. However, its mode of action is poorly understood. We determined a 2.6 Å resolution cryoelectron microscopy structure of a ternary complex between Escherichia coli topoisomerase DNA gyrase, a 217 bp double-stranded DNA fragment and albicidin. Albicidin employs a dual binding mechanism where one end of the molecule obstructs the crucial gyrase dimer interface, while the other intercalates between the fragments of cleaved DNA substrate. Thus, albicidin efficiently locks DNA gyrase, preventing it from religating DNA and completing its catalytic cycle. Two additional structures of this trapped state were determined using synthetic albicidin analogues that demonstrate improved solubility, and activity against a range of gyrase variants and E. coli topoisomerase IV. The extraordinary promiscuity of the DNA-intercalating region of albicidins and their excellent performance against fluoroquinolone-resistant bacteria holds great promise for the development of last-resort antibiotics.
- Malopolska Centre of Biotechnology, Jagiellonian University, Krakow, Poland.
Organizational Affiliation: