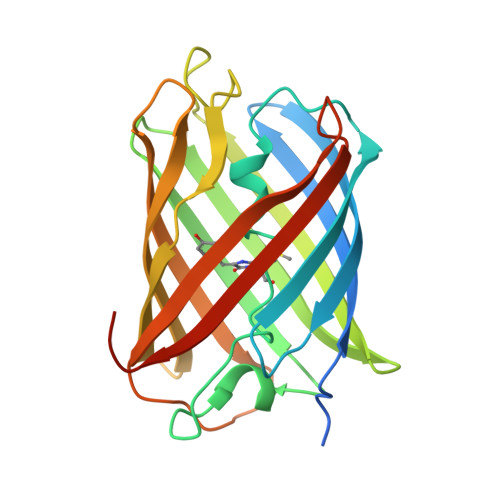
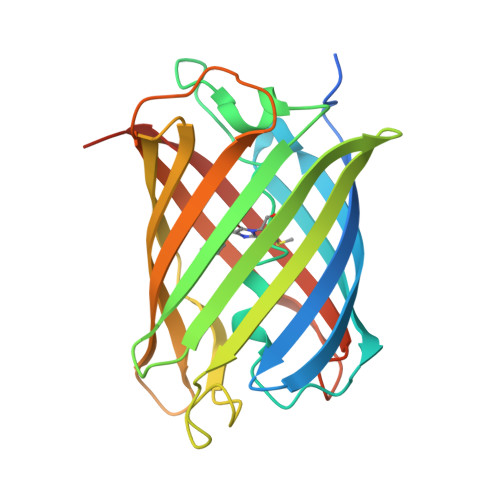
Generation of bright monomeric red fluorescent proteins via computational design of enhanced chromophore packing.
Legault, S., Fraser-Halberg, D.P., McAnelly, R.L., Eason, M.G., Thompson, M.C., Chica, R.A.(2022) Chem Sci 13: 1408-1418
- PubMed: 35222925
- DOI: https://doi.org/10.1039/d1sc05088e
- Primary Citation of Related Structures:
7RY2 - PubMed Abstract:
Red fluorescent proteins (RFPs) have found widespread application in chemical and biological research due to their longer emission wavelengths. Here, we use computational protein design to increase the quantum yield and thereby brightness of a dim monomeric RFP (mRojoA, quantum yield = 0.02) by optimizing chromophore packing with aliphatic residues, which we hypothesized would reduce torsional motions causing non-radiative decay. Experimental characterization of the top 10 designed sequences yielded mSandy1 ( λ em = 609 nm, quantum yield = 0.26), a variant with equivalent brightness to mCherry, a widely used RFP. We next used directed evolution to further increase brightness, resulting in mSandy2 ( λ em = 606 nm, quantum yield = 0.35), the brightest Discosoma sp. derived monomeric RFP with an emission maximum above 600 nm reported to date. Crystallographic analysis of mSandy2 showed that the chromophore p -hydroxybenzylidene moiety is sandwiched between the side chains of Leu63 and Ile197, a structural motif that has not previously been observed in RFPs, and confirms that aliphatic packing leads to chromophore rigidification. Our results demonstrate that computational protein design can be used to generate bright monomeric RFPs, which can serve as templates for the evolution of novel far-red fluorescent proteins.
- Department of Chemistry and Biomolecular Sciences, University of Ottawa 10 Marie-Curie Ottawa Ontario K1N 6N5 Canada rchica@uottawa.ca.
Organizational Affiliation: