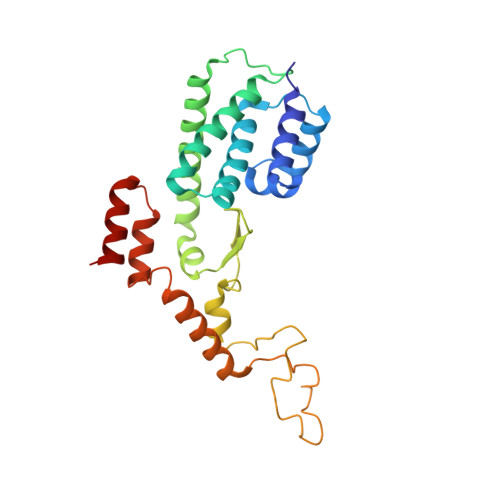
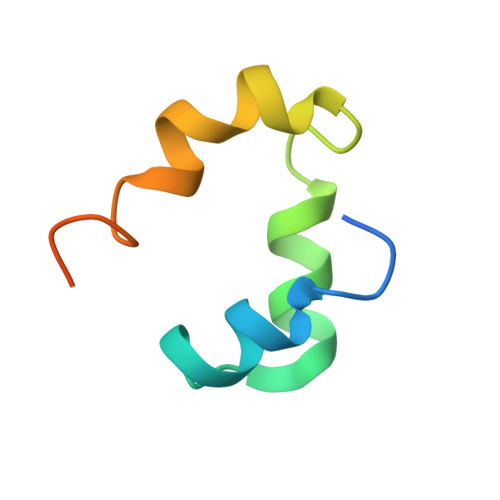
Non-canonical Staphylococcus aureus pathogenicity island repression.
Miguel-Romero, L., Alqasmi, M., Bacarizo, J., Tan, J.A., Cogdell, R.J., Chen, J., Byron, O., Christie, G.E., Marina, A., Penades, J.R.(2022) Nucleic Acids Res 50: 11109-11127
- PubMed: 36200825
- DOI: https://doi.org/10.1093/nar/gkac855
- Primary Citation of Related Structures:
7P4A, 7ZVI - PubMed Abstract:
Mobile genetic elements control their life cycles by the expression of a master repressor, whose function must be disabled to allow the spread of these elements in nature. Here, we describe an unprecedented repression-derepression mechanism involved in the transfer of Staphylococcus aureus pathogenicity islands (SaPIs). Contrary to the classical phage and SaPI repressors, which are dimers, the SaPI1 repressor StlSaPI1 presents a unique tetrameric conformation never seen before. Importantly, not just one but two tetramers are required for SaPI1 repression, which increases the novelty of the system. To derepress SaPI1, the phage-encoded protein Sri binds to and induces a conformational change in the DNA binding domains of StlSaPI1, preventing the binding of the repressor to its cognate StlSaPI1 sites. Finally, our findings demonstrate that this system is not exclusive to SaPI1 but widespread in nature. Overall, our results characterize a novel repression-induction system involved in the transfer of MGE-encoded virulence factors in nature.
- MRC Centre for Molecular Bacteriology and Infection, Imperial College London, SW7 2AZ, UK.
Organizational Affiliation: