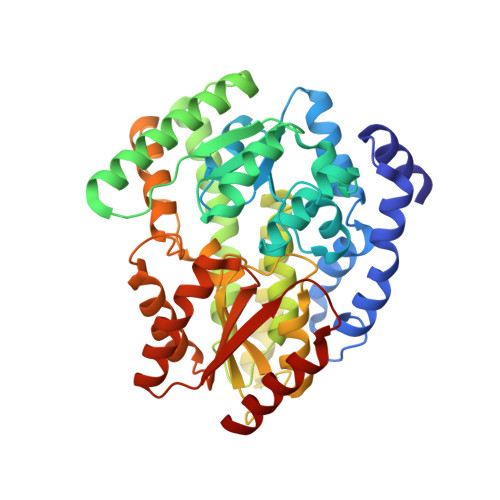
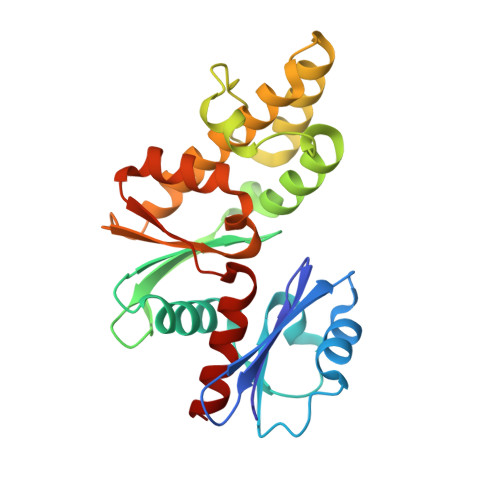
Structural basis for coupled ATP-driven electron transfer in the double-cubane cluster protein.
Jeoung, J.H., Nicklisch, S., Dobbek, H.(2022) Proc Natl Acad Sci U S A 119: e2203576119-e2203576119
- PubMed: 35905315
- DOI: https://doi.org/10.1073/pnas.2203576119
- Primary Citation of Related Structures:
7YYL, 7YZM, 7YZQ - PubMed Abstract:
Electron transfers coupled to the hydrolysis of ATP allow various metalloenzymes to catalyze reductions at very negative reduction potentials. The double-cubane cluster protein (DCCP) catalyzes the reduction of small molecules, such as acetylene and hydrazine, with electrons provided by its cognate ATP-hydrolyzing reductase (DCCP-R). How ATP-driven electron transfer occurs is not known. To resolve the structural basis for ATP-driven electron transfer, we solved the structures of the DCCP:DCCP-R complex in three different states. The structures show that the DCCP-R homodimer is covalently bridged by a [4Fe4S] cluster that is aligned with the twofold axis of the DCCP homodimer, positioning the [4Fe4S] cluster to enable electron transfer to both double-cubane clusters in the DCCP dimer. DCCP and DCCP-R form stable complexes independent of oxidation state or nucleotides present, and electron transfer requires the hydrolysis of ATP. Electron transfer appears to be additionally driven by modulating the angle between the helices binding the [4Fe4S] cluster. We observed hydrogen bond networks running from the ATP binding site via the [4Fe4S] cluster in DCCP-R to the double-cubane cluster in DCCP, allowing the propagation of conformational changes. Remarkable similarities between the DCCP:DCCP-R complex and the nonhomologous nitrogenases suggest a convergent evolution of catalytic strategies to achieve ATP-driven electron transfers between iron-sulfur clusters.
- Department of Biology, Humboldt-Universität zu Berlin, 10099 Berlin, Germany.
Organizational Affiliation: