Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site.
Lin, S., Du, T., Zhang, J., Wu, D., Tian, H., Zhang, K., Jiang, L., Lu, D., Sheng, L., Li, Y., Ji, M., Chen, X., Xu, H.(2022) J Med Chem 65: 16372-16391
- PubMed: 36511661
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01208
- Primary Citation of Related Structures:
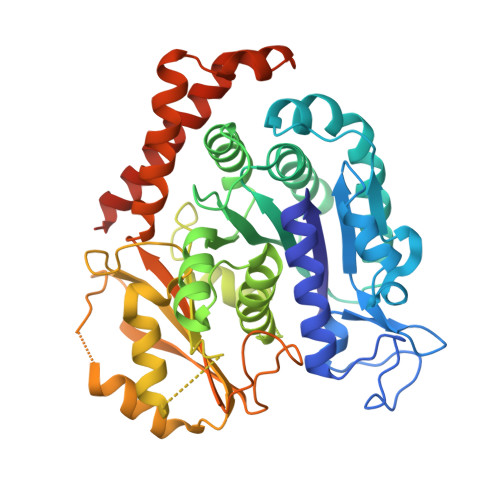
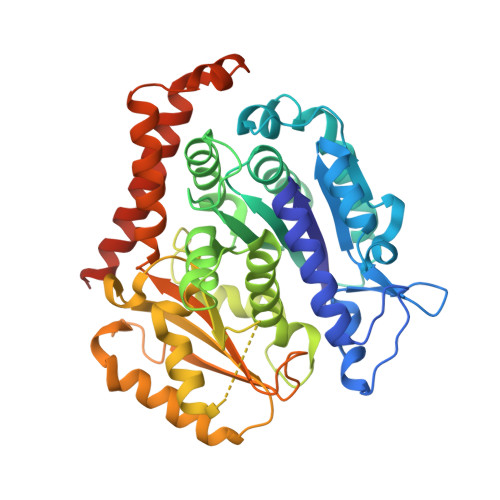
7YHN - PubMed Abstract:
Targeting the colchicine binding site on tubulin is a promising strategy to develop cancer therapeutics. Herein, we describe our systematic structure-activity relationship studies of benzamide derivatives that lead to an identification of a potent and orally active tubulin inhibitor 48 , which occupied all three zones of the colchicine binding site in the X-ray co-crystal structure, inhibited tubulin polymerization, promoted mitotic blockade and apoptosis, and exhibited significant antiproliferative activities against various cancer cell lines. Compound 48 demonstrated favorable pharmacokinetic profiles, robust in vivo antitumor efficacies, and acceptable safety profiles. Furthermore, 48 overcame drug resistance in the paclitaxel-resistant A549 xenograft model. Collectively, 48 has been advanced into further preclinical evaluation for the development of next-generation microtubule-targeting drugs.
- State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China.
Organizational Affiliation: