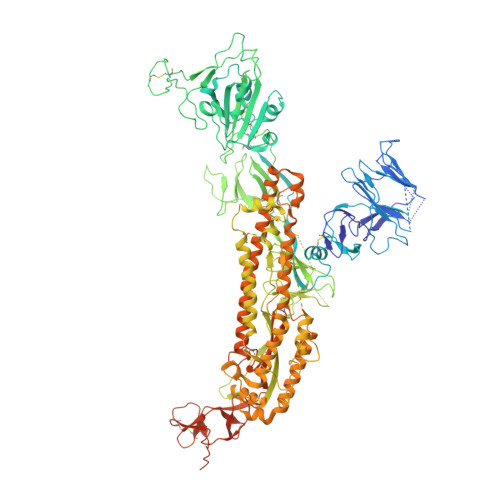
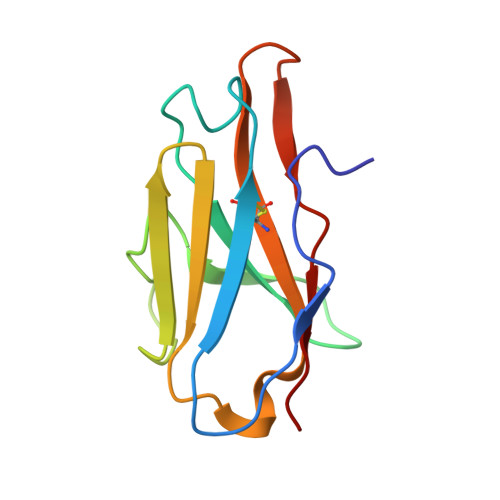
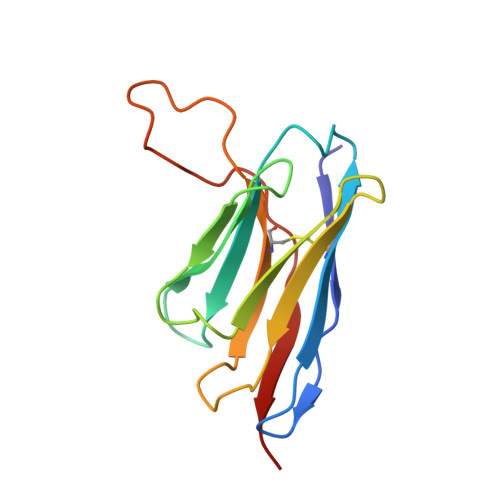
Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Moriyama, S., Anraku, Y., Taminishi, S., Adachi, Y., Kuroda, D., Kita, S., Higuchi, Y., Kirita, Y., Kotaki, R., Tonouchi, K., Yumoto, K., Suzuki, T., Someya, T., Fukuhara, H., Kuroda, Y., Yamamoto, T., Onodera, T., Fukushi, S., Maeda, K., Nakamura-Uchiyama, F., Hashiguchi, T., Hoshino, A., Maenaka, K., Takahashi, Y.(2023) Nat Commun 14: 4198-4198
- PubMed: 37452031
- DOI: https://doi.org/10.1038/s41467-023-39890-8
- Primary Citation of Related Structures:
7YH6, 7YH7, 8HES, 8HGL, 8HGM - PubMed Abstract:
SARS-CoV-2 Omicron subvariants have evolved to evade receptor-binding site (RBS) antibodies that exist in diverse individuals as public antibody clones. We rationally selected RBS antibodies resilient to mutations in emerging Omicron subvariants. Y489 was identified as a site of virus vulnerability and a common footprint of broadly neutralizing antibodies against the subvariants. Multiple Y489-binding antibodies were encoded by public clonotypes and additionally recognized F486, potentially accounting for the emergence of Omicron subvariants harboring the F486V mutation. However, a subclass of antibodies broadly neutralized BA.4/BA.5 variants via hydrophobic binding sites of rare clonotypes along with high mutation-resilience under escape mutation screening. A computationally designed antibody based on one of the Y489-binding antibodies, NIV-10/FD03, was able to bind XBB with any 486 mutation and neutralized XBB.1.5. The structural basis for the mutation-resilience of this Y489-binding antibody group may provide important insights into the design of therapeutics resistant to viral escape.
- Research Center for Drug and Vaccine Development, National Institute of Infectious Diseases; Shinjuku-ku, Tokyo, 162-8640, Japan. sayamrym@niid.go.jp.
Organizational Affiliation: