Structural insight into an anti-BRIL Fab as a G-protein-coupled receptor crystallization chaperone.
Miyagi, H., Suzuki, M., Yasunaga, M., Asada, H., Iwata, S., Saito, J.I.(2023) Acta Crystallogr D Struct Biol 79: 435-441
- PubMed: 37098908
- DOI: https://doi.org/10.1107/S205979832300311X
- Primary Citation of Related Structures:
7XRZ - PubMed Abstract:
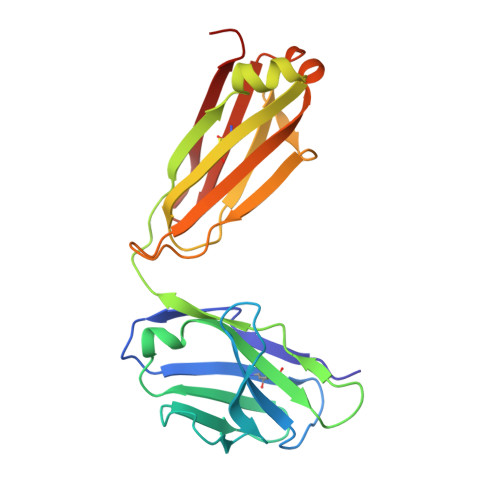
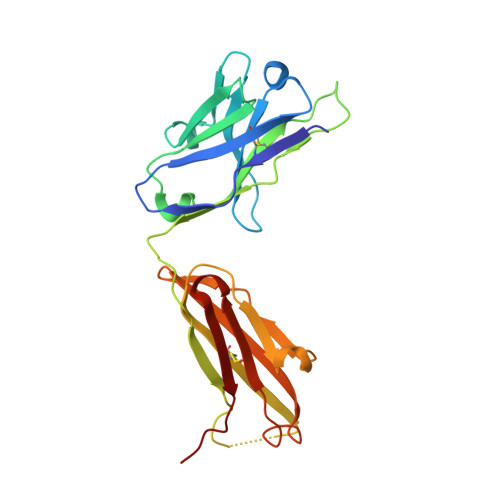
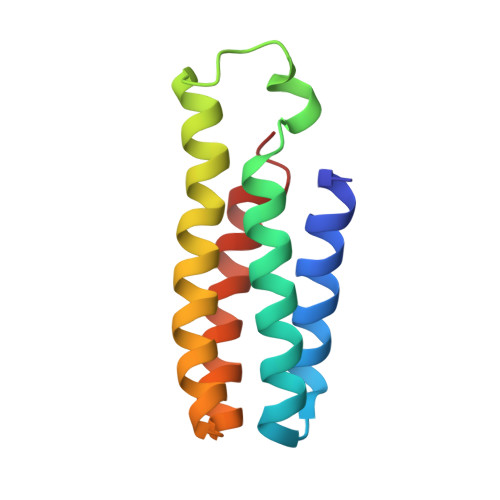
Structure determination of G-protein-coupled receptors (GPCRs) is key for the successful development of efficient drugs targeting GPCRs. BRIL is a thermostabilized apocytochrome b 562 (with M7W/H102I/R106L mutations) from Escherichia coli and is often used as a GPCR fusion protein for expression and crystallization. SRP2070Fab, an anti-BRIL antibody Fab fragment, has been reported to facilitate and enhance the crystallization of BRIL-fused GPCRs as a crystallization chaperone. This study was conducted to characterize the high-resolution crystal structure of the BRIL-SRP2070Fab complex. The structure of the BRIL-SRP2070Fab complex was determined at 2.1 Å resolution. This high-resolution structure elucidates the binding interaction between BRIL and SRP2070Fab. When binding to BRIL, SRP2070Fab recognizes conformational epitopes, not linear epitopes, on the surface of BRIL helices III and IV, thereby binding perpendicularly to the helices, which indicates stable binding. Additionally, the packing contacts of the BRIL-SRP2070Fab co-crystal are largely due to SRP2070Fab rather than BRIL. The accumulation of SRP2070Fab molecules by stacking is remarkable and is consistent with the finding that stacking of SRP2070Fab is predominant in known crystal structures of BRIL-fused GPCRs complexed with SRP2070Fab. These findings clarified the mechanism of SRP2070Fab as a crystallization chaperone. Moreover, these data will be useful in the structure-based drug design of membrane-protein drug targets.
- R&D Division, Kyowa Kirin Co. Ltd, Tokyo, Japan.
Organizational Affiliation: