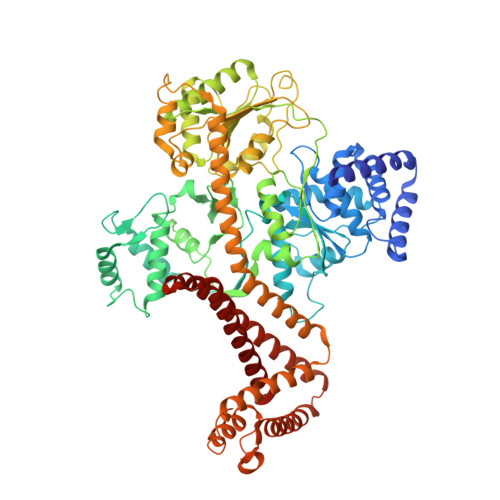
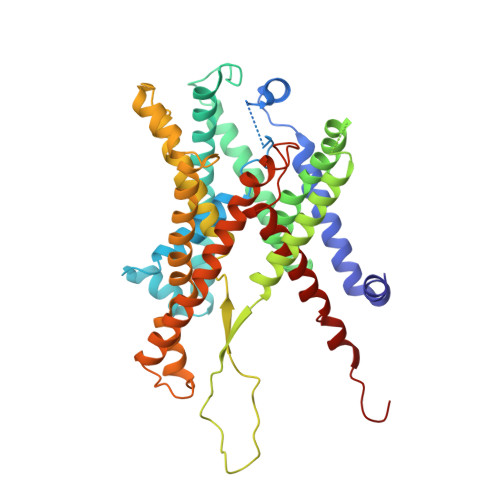
Structural basis of SecA-mediated protein translocation.
Dong, L., Yang, S., Chen, J., Wu, X., Sun, D., Song, C., Li, L.(2023) Proc Natl Acad Sci U S A 120: e2208070120-e2208070120
- PubMed: 36598944
- DOI: https://doi.org/10.1073/pnas.2208070120
- Primary Citation of Related Structures:
7XHA, 7XHB - PubMed Abstract:
Secretory proteins are cotranslationally or posttranslationally translocated across lipid membranes via a protein-conducting channel named SecY in prokaryotes and Sec61 in eukaryotes. The vast majority of secretory proteins in bacteria are driven through the channel posttranslationally by SecA, a highly conserved ATPase. How a polypeptide chain is moved by SecA through the SecY channel is poorly understood. Here, we report electron cryomicroscopy structures of the active SecA-SecY translocon with a polypeptide substrate. The substrate is captured in different translocation states when clamped by SecA with different nucleotides. Upon binding of an ATP analog, SecA undergoes global conformational changes to push the polypeptide substrate toward the channel in a way similar to how the RecA-like helicases translocate their nucleic acid substrates. The movements of the polypeptide substrates in the SecA-SecY translocon share a similar structural basis to those in the ribosome-SecY complex during cotranslational translocation.
- State Key Laboratory of Membrane Biology, Peking-Tsinghua Center for Life Sciences, School of Life Sciences, Peking University, Beijing 100871, China.
Organizational Affiliation: