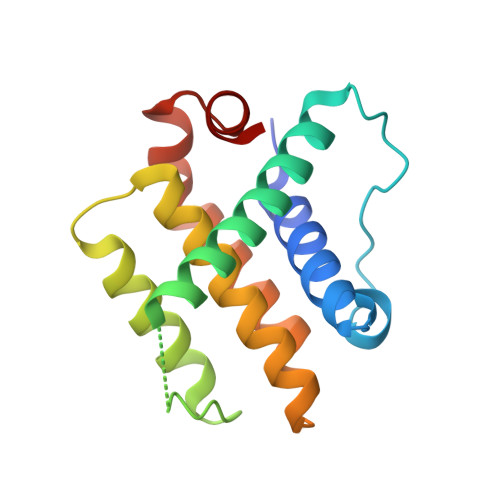
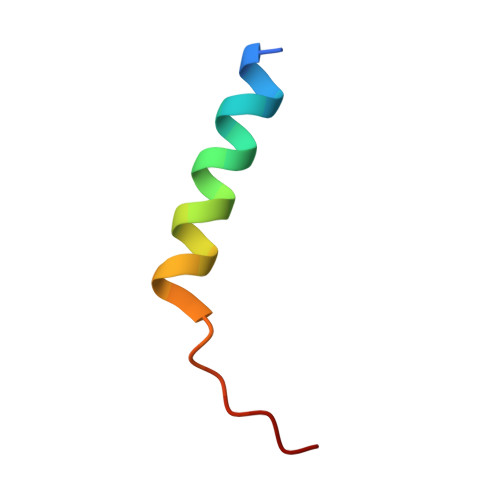
Structural and biochemical analyses of Bcl-xL in complex with the BH3 domain of peroxisomal testis-specific 1.
Lim, D., Jin, S., Shin, H.C., Kim, W., Choi, J.S., Oh, D.B., Kim, S.J., Seo, J., Ku, B.(2022) Biochem Biophys Res Commun 625: 174-180
- PubMed: 35964379
- DOI: https://doi.org/10.1016/j.bbrc.2022.08.009
- Primary Citation of Related Structures:
7WJH - PubMed Abstract:
Antiapoptotic B-cell lymphoma-2 (Bcl-2) proteins suppress apoptosis by interacting with proapoptotic regulators. They commonly contain a hydrophobic groove where the Bcl-2 homology 3 (BH3) domain of Bcl-2 family members or BH3 domain-containing non-Bcl-2 family proteins can be accommodated. Peroxisomal testis-specific 1 (Pxt1) was previously identified as a male germ cell-specific protein whose overexpression causes germ cell apoptosis and infertility in male mice. Sequence and biochemical analyses also showed that human Pxt1, which is composed of 134 amino acids and is longer than mouse Pxt1 consisting of only 51 amino acids, has a BH3 domain that interacts with antiapoptotic Bcl-2 proteins, including Bcl-2 and Bcl-xL. In this study, we determined the crystal structure of Bcl-xL bound to the human Pxt1 BH3 domain. The five BH3 consensus residues are well conserved in the human Pxt1 BH3 domain and make a critical contribution to the complex formation in a canonical manner. Structural and biochemical analyses also demonstrated that Bcl-xL interacts with the BH3 domain of human Pxt1 but not with that of mouse Pxt1, and that residues 76-83 of human Pxt1, absent in mouse Pxt1, play a pivotal role in the intermolecular binding to Bcl-xL. While Bcl-xL consistently colocalized with human Pxt1 in mitochondria, it did not do so with mouse Pxt1, when expressed in HeLa cells. Collectively, these data verified that human and mouse Pxt1 differ in their binding ability to the antiapoptotic regulator Bcl-xL, which might affect their functionality in controlling apoptosis.
- Critical Diseases Diagnostics Convergence Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, 34141, South Korea; Department of Biochemistry, Chungnam National University, Daejeon, 34134, South Korea.
Organizational Affiliation: