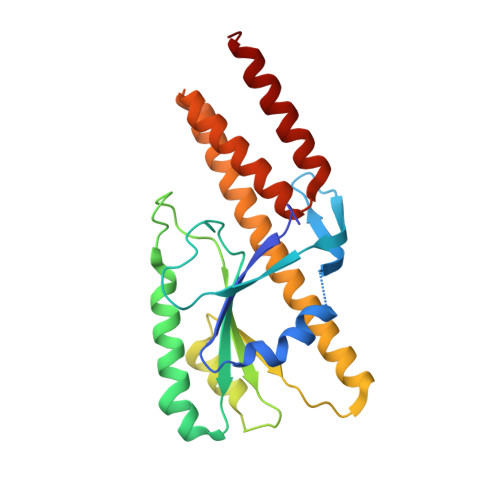
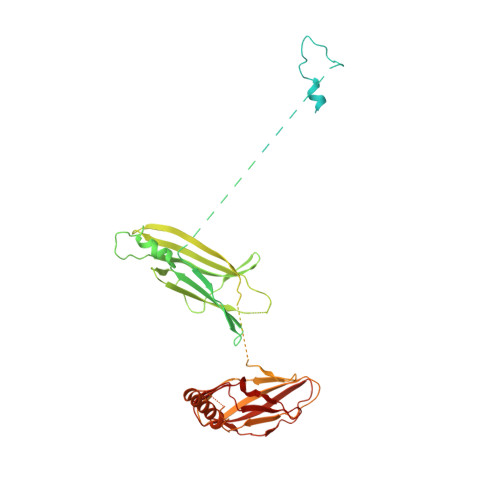
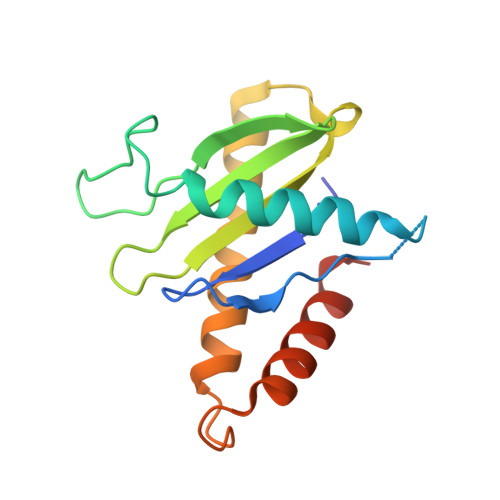
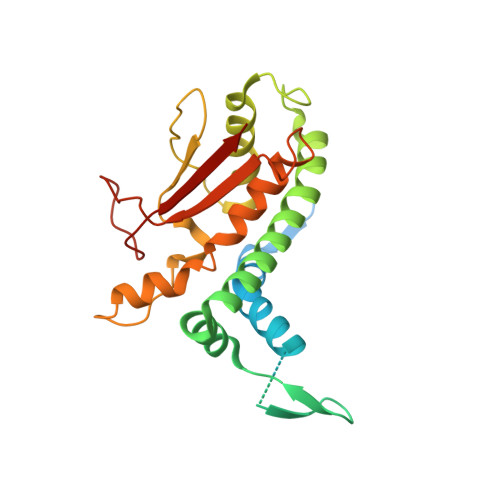
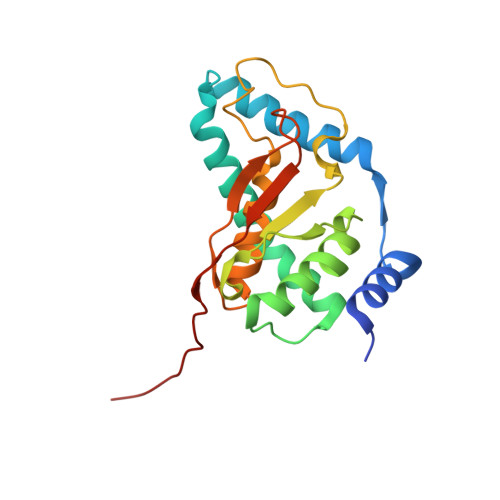
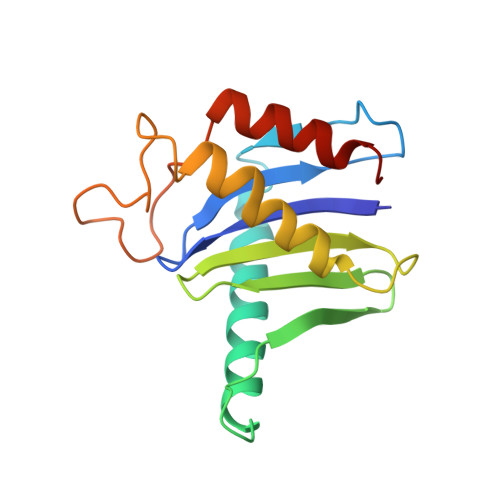
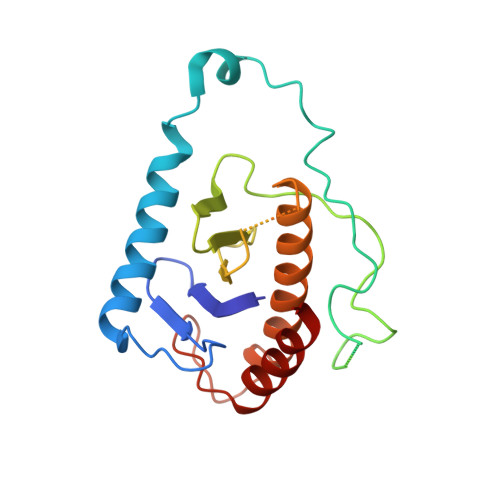
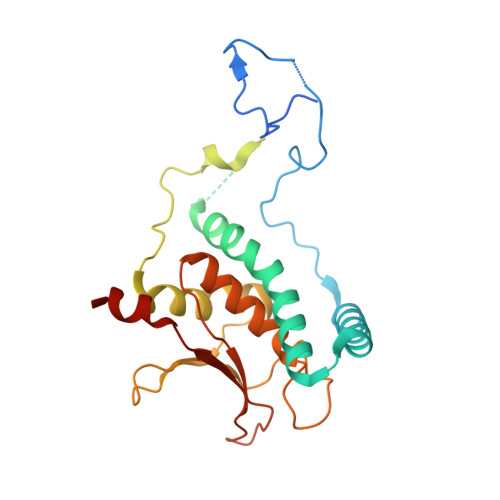
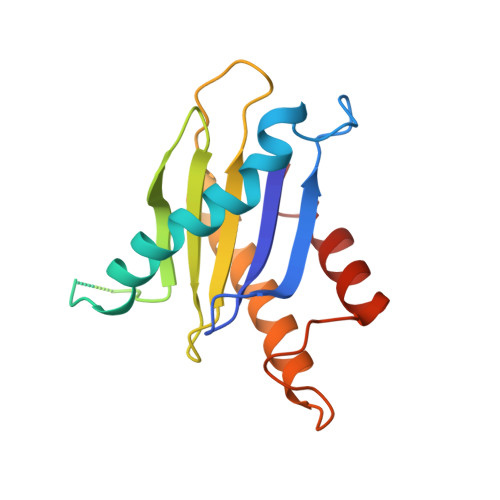
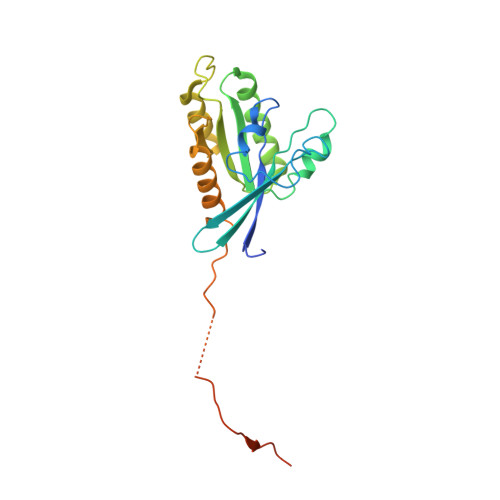
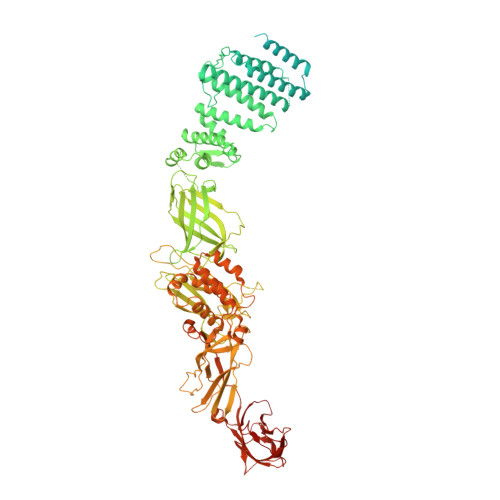
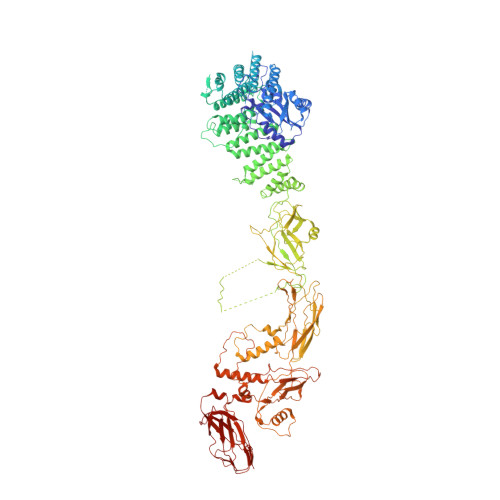
Structure of a TRAPPII-Rab11 activation intermediate reveals GTPase substrate selection mechanisms.
Bagde, S.R., Fromme, J.C.(2022) Sci Adv 8: eabn7446-eabn7446
- PubMed: 35559680
- DOI: https://doi.org/10.1126/sciadv.abn7446
- Primary Citation of Related Structures:
7U05, 7U06 - PubMed Abstract:
Rab1 and Rab11 are essential regulators of the eukaryotic secretory and endocytic recycling pathways. The transport protein particle (TRAPP) complexes activate these guanosine triphosphatases via nucleotide exchange using a shared set of core subunits. The basal specificity of the TRAPP core is toward Rab1, yet the TRAPPII complex is specific for Rab11. A steric gating mechanism has been proposed to explain TRAPPII counterselection against Rab1. Here, we present cryo-electron microscopy structures of the 22-subunit TRAPPII complex from budding yeast, including a TRAPPII-Rab11 nucleotide exchange intermediate. The Trs130 subunit provides a "leg" that positions the active site distal to the membrane surface, and this leg is required for steric gating. The related TRAPPIII complex is unable to activate Rab11 because of a repulsive interaction, which TRAPPII surmounts using the Trs120 subunit as a "lid" to enclose the active site. TRAPPII also adopts an open conformation enabling Rab11 to access and exit from the active site chamber.
- Department of Molecular Biology and Genetics and Weill Institute for Cell and Molecular Biology, Cornell University, Ithaca, NY 14853, USA.
Organizational Affiliation: