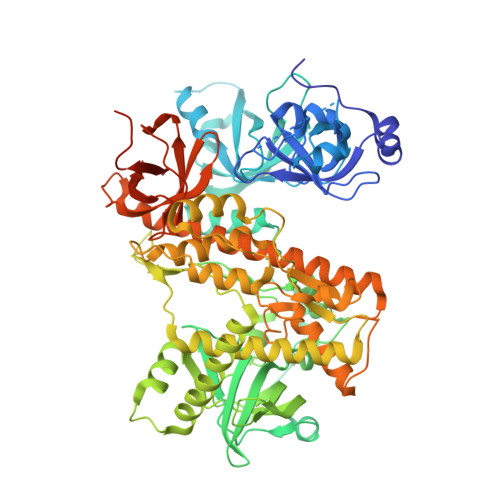
Trypanosoma brucei RRP44: a versatile enzyme for processing structured and non-structured RNA substrates.
Cesaro, G., da Soler, H.T., Guerra-Slompo, E.P., Haouz, A., Legrand, P., Zanchin, N.I.T., Guimaraes, B.G.(2023) Nucleic Acids Res 51: 380-395
- PubMed: 36583334
- DOI: https://doi.org/10.1093/nar/gkac1199
- Primary Citation of Related Structures:
7TUV - PubMed Abstract:
Rrp44/Dis3 is a conserved eukaryotic ribonuclease that acts on processing and degradation of nearly all types of RNA. It contains an endo- (PIN) and an exonucleolytic (RNB) domain and, its depletion in model organisms supports its essential function for cell viability. In Trypanosoma brucei, depletion of Rrp44 (TbRRP44) blocks maturation of ribosomal RNA, leading to disruption of ribosome synthesis and inhibition of cell proliferation. We have determined the crystal structure of the exoribonucleolytic module of TbRRP44 in an active conformation, revealing novel details of the catalytic mechanism of the RNB domain. For the first time, the position of the second magnesium involved in the two-metal-ion mechanism was determined for a member of the RNase II family. In vitro, TbRRP44 acts preferentially on non-structured uridine-rich RNA substrates. However, we demonstrated for the first time that both TbRRP44 and its homologue from Saccharomyces cerevisiae can also degrade structured substrates without 3'-end overhang, suggesting that Rrp44/Dis3 ribonucleases may be involved in degradation of a wider panel of RNA than has been assumed. Interestingly, deletion of TbRRP44 PIN domain impairs RNA binding to different extents, depending on the type of substrate.
- Carlos Chagas Institute, Oswaldo Cruz Foundation, FIOCRUZ, Curitiba-PR, Brazil.
Organizational Affiliation: