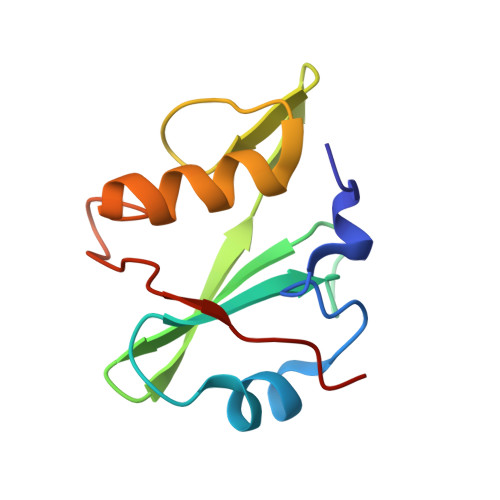
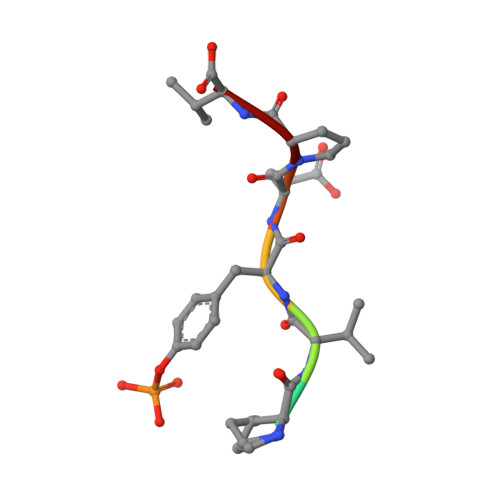
Engineered SH2 Domains for Targeted Phosphoproteomics.
Martyn, G.D., Veggiani, G., Kusebauch, U., Morrone, S.R., Yates, B.P., Singer, A.U., Tong, J., Manczyk, N., Gish, G., Sun, Z., Kurinov, I., Sicheri, F., Moran, M.F., Moritz, R.L., Sidhu, S.S.(2022) ACS Chem Biol 17: 1472-1484
- PubMed: 35613471
- DOI: https://doi.org/10.1021/acschembio.2c00051
- Primary Citation of Related Structures:
7T1K, 7T1L, 7T1U - PubMed Abstract:
A comprehensive analysis of the phosphoproteome is essential for understanding molecular mechanisms of human diseases. However, current tools used to enrich phosphotyrosine (pTyr) are limited in their applicability and scope. Here, we engineered new superbinder Src-Homology 2 (SH2) domains that enrich diverse sets of pTyr-peptides. We used phage display to select a Fes-SH2 domain variant (superFes; sFes 1 ) with high affinity for pTyr and solved its structure bound to a pTyr-peptide. We performed systematic structure-function analyses of the superbinding mechanisms of sFes 1 and superSrc-SH2 (sSrc 1 ), another SH2 superbinder. We grafted the superbinder motifs from sFes 1 and sSrc 1 into 17 additional SH2 domains and confirmed increased binding affinity for specific pTyr-peptides. Using mass spectrometry (MS), we demonstrated that SH2 superbinders have distinct specificity profiles and superior capabilities to enrich pTyr-peptides. Finally, using combinations of SH2 superbinders as affinity purification (AP) tools we showed that unique subsets of pTyr-peptides can be enriched with unparalleled depth and coverage.
- Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, 160 College Street, Toronto, Ontario M5S3E1, Canada.
Organizational Affiliation: