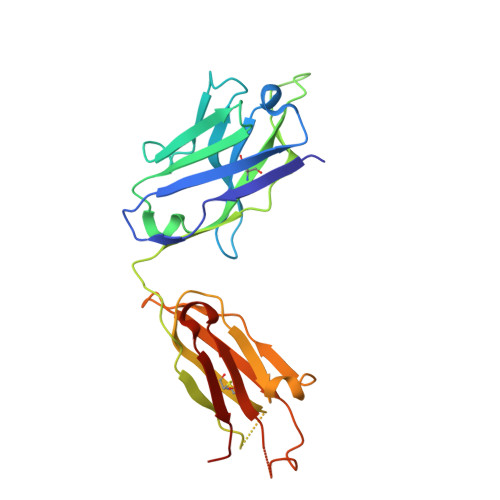
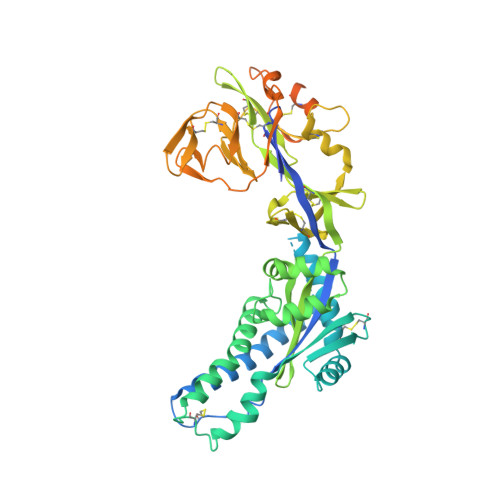
Structure-based design of prefusion-stabilized human metapneumovirus fusion proteins.
Hsieh, C.L., Rush, S.A., Palomo, C., Chou, C.W., Pickens, W., Mas, V., McLellan, J.S.(2022) Nat Commun 13: 1299-1299
- PubMed: 35288548
- DOI: https://doi.org/10.1038/s41467-022-28931-3
- Primary Citation of Related Structures:
7SEJ, 7SEM - PubMed Abstract:
The human metapneumovirus (hMPV) fusion (F) protein is essential for viral entry and is a key target of neutralizing antibodies and vaccine development. The prefusion conformation is thought to be the optimal vaccine antigen, but previously described prefusion F proteins expressed poorly and were not well stabilized. Here, we use structures of hMPV F to guide the design of 42 variants containing stabilizing substitutions. Through combinatorial addition of disulfide bonds, cavity-filling substitutions, and improved electrostatic interactions, we describe a prefusion-stabilized F protein (DS-CavEs2) that expresses at 15 mg/L and has a melting temperature of 71.9 °C. Crystal structures of two prefusion-stabilized hMPV F variants reveal that antigenic surfaces are largely unperturbed. Importantly, immunization of mice with DS-CavEs2 elicits significantly higher neutralizing antibody titers against hMPV A1 and B1 viruses than postfusion F. The improved properties of DS-CavEs2 will advance the development of hMPV vaccines and the isolation of therapeutic antibodies.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX, USA.
Organizational Affiliation: