Antibody blockade of CD96 by distinct molecular mechanisms.
Lee, P.S., Chau, B., Barman, I., Bee, C., Jashnani, A., Hogan, J.M., Aguilar, B., Dollinger, G., Rajpal, A., Strop, P.(2021) MAbs 13: 1979800-1979800
- PubMed: 34595996
- DOI: https://doi.org/10.1080/19420862.2021.1979800
- Primary Citation of Related Structures:
7S11, 7S13 - PubMed Abstract:
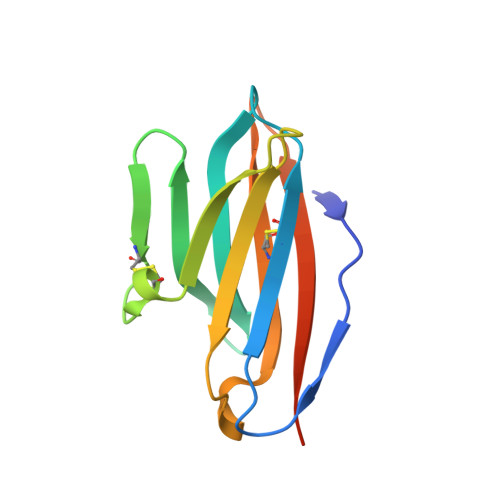
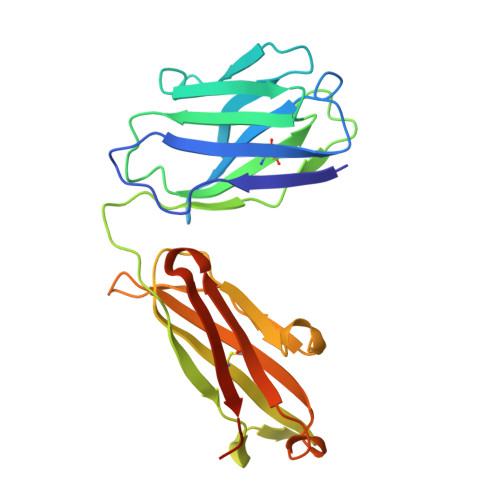
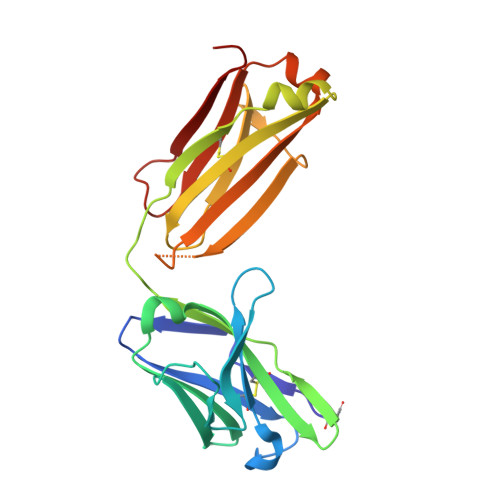
The molecular interactions of mouse CD96 to CD155 ligand and to two surrogate antibodies have been investigated. Biophysical and structural studies demonstrate that CD96 forms a homodimer but assembles as 1:1 heterodimeric complexes with CD155 or with one of the surrogate antibodies, which compete for the same binding interface. In comparison, the other surrogate antibody binds across the mouse CD96 dimer and recognizes a quaternary epitope spanning both protomers to block exposure of the ligand-binding site. This study reveals different blocking mechanisms and modalities of these two antibodies and may provide insight into the functional effects of antibodies against CD96.
- Discovery Biotherapeutics, Bristol Myers Squibb, Redwood City, CA, USA.
Organizational Affiliation: