Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Arellano-Santoyo, H., Hernandez-Lopez, R.A., Stokasimov, E., Wang, R.Y.-R., Pellman, D., Lesczhiner, A.E.(2024) bioRxiv
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
(2024) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin alpha-1B chain | 451 | Sus scrofa | Mutation(s): 0 EC: 3.6.5 | 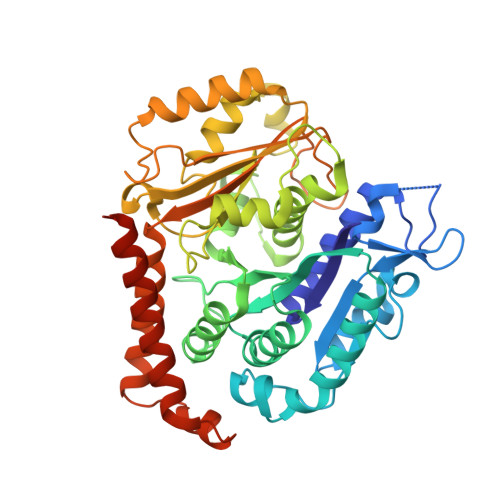 | |
UniProt | |||||
Find proteins for Q2XVP4 (Sus scrofa) Explore Q2XVP4 Go to UniProtKB: Q2XVP4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2XVP4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin beta chain | 445 | Sus scrofa | Mutation(s): 0 | 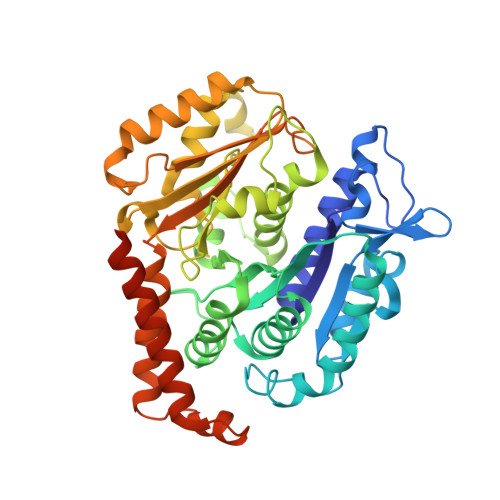 | |
UniProt | |||||
Find proteins for P02554 (Sus scrofa) Explore P02554 Go to UniProtKB: P02554 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02554 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| kinesin-8/ Kip3 | 355 | Saccharomyces cerevisiae | Mutation(s): 0 | 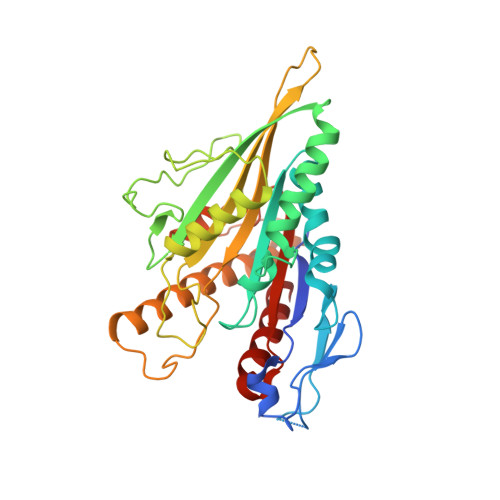 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP (Subject of Investigation/LOI) Query on GTP | BA [auth A] FB [auth L] HA [auth C] LB [auth N] NA [auth E] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| G2P (Subject of Investigation/LOI) Query on G2P | BB [auth J] DA [auth B] HB [auth M] JA [auth D] NB [auth O] | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER C11 H18 N5 O13 P3 GXTIEXDFEKYVGY-KQYNXXCUSA-N |  | ||
| ANP (Subject of Investigation/LOI) Query on ANP | BC [auth h] DB [auth d] FA [auth K] JB [auth e] LA [auth a] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | AB [auth I] AC [auth S] CA [auth A] CB [auth J] CC [auth h] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | Rosetta | |
| RECONSTRUCTION | FREALIGN |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |