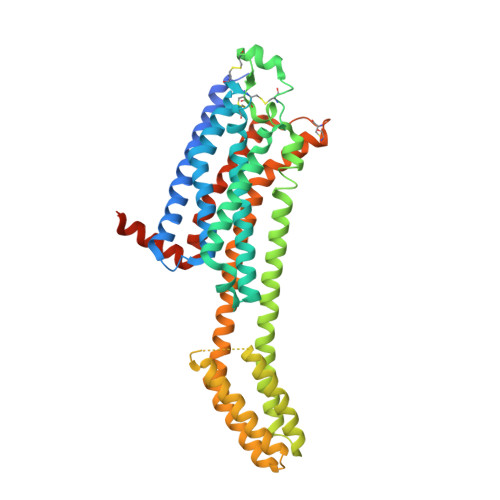
MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Martynowycz, M.W., Shiriaeva, A., Ge, X., Hattne, J., Nannenga, B.L., Cherezov, V., Gonen, T.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34462357
- DOI: https://doi.org/10.1073/pnas.2106041118
- Primary Citation of Related Structures:
7RM5 - PubMed Abstract:
G protein-coupled receptors (GPCRs), or seven-transmembrane receptors, are a superfamily of membrane proteins that are critically important to physiological processes in the human body. Determining high-resolution structures of GPCRs without bound cognate signaling partners, such as a G protein, requires crystallization in lipidic cubic phase (LCP). GPCR crystals grown in LCP are often too small for traditional X-ray crystallography. These microcrystals are ideal for investigation by microcrystal electron diffraction (MicroED), but the gel-like nature of LCP makes traditional approaches to MicroED sample preparation insurmountable. Here, we show that the structure of a human A 2A adenosine receptor can be determined by MicroED after converting the LCP into the sponge phase followed by focused ion-beam milling. We determined the structure of the A 2A adenosine receptor to 2.8-Å resolution and resolved an antagonist in its orthosteric ligand-binding site, as well as four cholesterol molecules bound around the receptor. This study lays the groundwork for future structural studies of lipid-embedded membrane proteins by MicroED using single microcrystals that would be impossible with other crystallographic methods.
- HHMI, University of California, Los Angeles, CA 90095.
Organizational Affiliation: