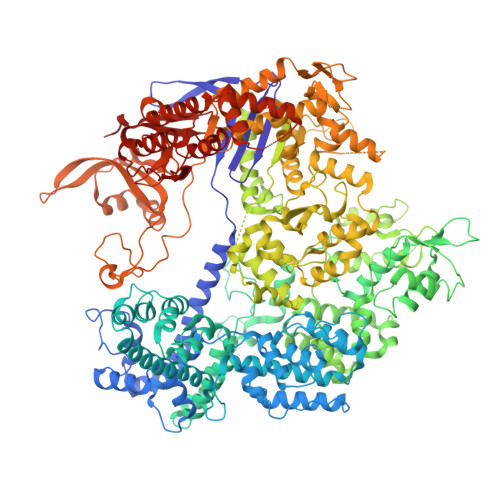
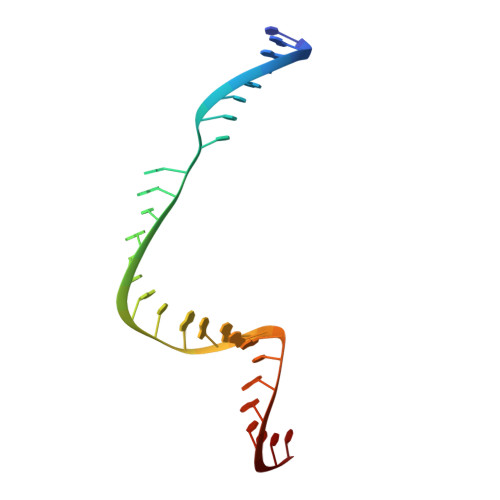
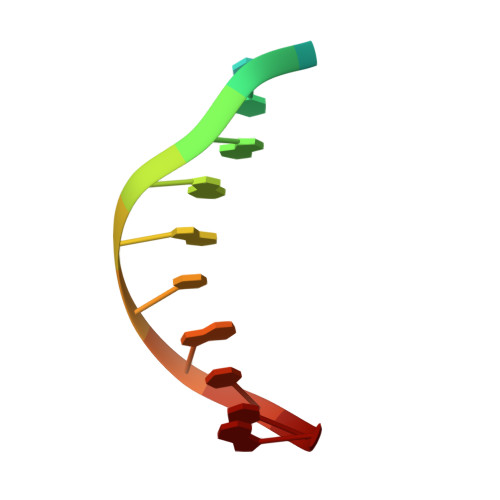
Structural basis for Cas9 off-target activity.
Pacesa, M., Lin, C.H., Clery, A., Saha, A., Arantes, P.R., Bargsten, K., Irby, M.J., Allain, F.H., Palermo, G., Cameron, P., Donohoue, P.D., Jinek, M.(2022) Cell 185: 4067-4081.e21
- PubMed: 36306733
- DOI: https://doi.org/10.1016/j.cell.2022.09.026
- Primary Citation of Related Structures:
7QQO, 7QQP, 7QQQ, 7QQR, 7QQS, 7QQT, 7QQU, 7QQV, 7QQW, 7QQX, 7QQZ, 7QR0, 7QR1, 7QR5, 7QR7, 7QR8, 7ZO1 - PubMed Abstract:
The target DNA specificity of the CRISPR-associated genome editor nuclease Cas9 is determined by complementarity to a 20-nucleotide segment in its guide RNA. However, Cas9 can bind and cleave partially complementary off-target sequences, which raises safety concerns for its use in clinical applications. Here, we report crystallographic structures of Cas9 bound to bona fide off-target substrates, revealing that off-target binding is enabled by a range of noncanonical base-pairing interactions within the guide:off-target heteroduplex. Off-target substrates containing single-nucleotide deletions relative to the guide RNA are accommodated by base skipping or multiple noncanonical base pairs rather than RNA bulge formation. Finally, PAM-distal mismatches result in duplex unpairing and induce a conformational change in the Cas9 REC lobe that perturbs its conformational activation. Together, these insights provide a structural rationale for the off-target activity of Cas9 and contribute to the improved rational design of guide RNAs and off-target prediction algorithms.
- Department of Biochemistry, University of Zurich, Winterthurerstrasse 190, 8057 Zurich, Switzerland.
Organizational Affiliation: