Structure of full length SpoT
Garcia-Pino, A., Tamman, H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ACT domain protein | 701 | Acinetobacter baumannii | Mutation(s): 0 Gene Names: spoT_1, relA_1, relA_3, spoT, A7M90_14410, AB945B12_01945, Aba9201_15290, ABCAM1_3401, ABKPCSM17A_01292, ABR2091_3374... EC: 2.7.6.5 (PDB Primary Data), 3.1.7.2 (UniProt) | 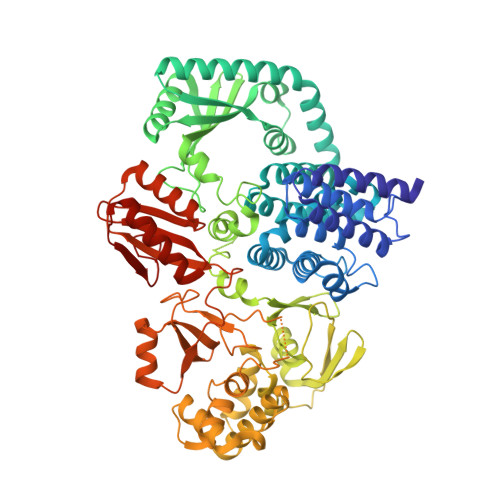 | |
UniProt | |||||
Find proteins for V5V8V7 (Acinetobacter baumannii) Explore V5V8V7 Go to UniProtKB: V5V8V7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | V5V8V7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| G4P (Subject of Investigation/LOI) Query on G4P | G [auth A], M [auth B], Q [auth C], Y [auth D] | GUANOSINE-5',3'-TETRAPHOSPHATE C10 H17 N5 O17 P4 BUFLLCUFNHESEH-UUOKFMHZSA-N |  | ||
| FLC Query on FLC | H [auth A], R [auth C] | CITRATE ANION C6 H5 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-K |  | ||
| GOL Query on GOL | I [auth A], N [auth B], S [auth C], T [auth C] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | F [auth A], L [auth B], P [auth C], X [auth D] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MN (Subject of Investigation/LOI) Query on MN | E [auth A], K [auth B], O [auth C], W [auth D] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| CL Query on CL | AA [auth D], J [auth A], V [auth C] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | U [auth C], Z [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 128.791 | α = 90 |
| b = 133.761 | β = 90 |
| c = 211.328 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |