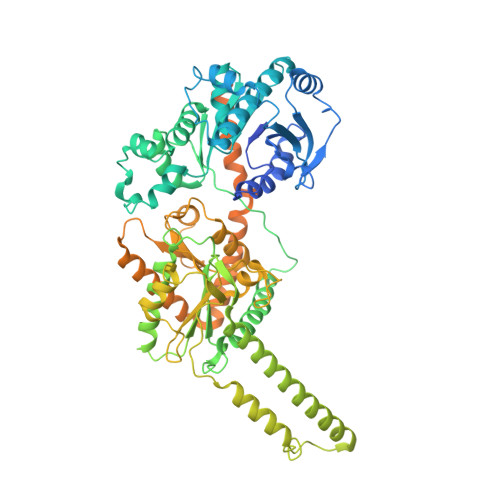
Molecular basis for the regulation of human glycogen synthase by phosphorylation and glucose-6-phosphate.
McCorvie, T.J., Loria, P.M., Tu, M., Han, S., Shrestha, L., Froese, D.S., Ferreira, I.M., Berg, A.P., Yue, W.W.(2022) Nat Struct Mol Biol 29: 628-638
- PubMed: 35835870
- DOI: https://doi.org/10.1038/s41594-022-00799-3
- Primary Citation of Related Structures:
7Q0B, 7Q0S, 7Q12, 7Q13 - PubMed Abstract:
Glycogen synthase (GYS1) is the central enzyme in muscle glycogen biosynthesis. GYS1 activity is inhibited by phosphorylation of its amino (N) and carboxyl (C) termini, which is relieved by allosteric activation of glucose-6-phosphate (Glc6P). We present cryo-EM structures at 3.0-4.0 Å resolution of phosphorylated human GYS1, in complex with a minimal interacting region of glycogenin, in the inhibited, activated and catalytically competent states. Phosphorylations of specific terminal residues are sensed by different arginine clusters, locking the GYS1 tetramer in an inhibited state via intersubunit interactions. The Glc6P activator promotes conformational change by disrupting these interactions and increases the flexibility of GYS1, such that it is poised to adopt a catalytically competent state when the sugar donor UDP-glucose (UDP-glc) binds. We also identify an inhibited-like conformation that has not transitioned into the activated state, in which the locking interaction of phosphorylation with the arginine cluster impedes subsequent conformational changes due to Glc6P binding. Our results address longstanding questions regarding the mechanism of human GYS1 regulation.
- Centre for Medicines Discovery, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, UK.
Organizational Affiliation: