A novel Pyk2-derived peptide inhibits invadopodia-mediated breast cancer metastasis.
Twafra, S., Sokolik, C.G., Sneh, T., Srikanth, K.D., Meirson, T., Genna, A., Chill, J.H., Gil-Henn, H.(2023) Oncogene 42: 278-292
- PubMed: 36258022
- DOI: https://doi.org/10.1038/s41388-022-02481-w
- Primary Citation of Related Structures:
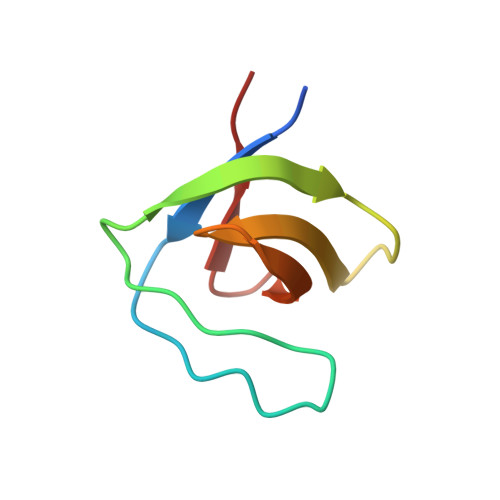
7PLL - PubMed Abstract:
Dissemination of cancer cells from the primary tumor into distant body tissues and organs is the leading cause of death in cancer patients. While most clinical strategies aim to reduce or impede the growth of the primary tumor, no treatment to eradicate metastatic cancer exists at present. Metastasis is mediated by feet-like cytoskeletal structures called invadopodia which allow cells to penetrate through the basement membrane and intravasate into blood vessels during their spread to distant tissues and organs. The non-receptor tyrosine kinase Pyk2 is highly expressed in breast cancer, where it mediates invadopodia formation and function via interaction with the actin-nucleation-promoting factor cortactin. Here, we designed a cell-permeable peptide inhibitor that contains the second proline-rich region (PRR2) sequence of Pyk2, which binds to the SH3 domain of cortactin and inhibits the interaction between Pyk2 and cortactin in invadopodia. The Pyk2-PRR2 peptide blocks spontaneous lung metastasis in immune-competent mice by inhibiting cortactin tyrosine phosphorylation and actin polymerization-mediated maturation and activation of invadopodia, leading to reduced MMP-dependent tumor cell invasiveness. The native structure of the Pyk2-PRR2:cortactin-SH3 complex was determined using nuclear magnetic resonance (NMR), revealing an extended class II interaction surface spanning the canonical binding groove and a second hydrophobic surface which significantly contributes to ligand affinity. Using structure-guided design, we created a mutant peptide lacking critical residues involved in binding that failed to inhibit invadopodia maturation and function and consequent metastatic dissemination in mice. Our findings shed light on the specific molecular interactions between Pyk2 and cortactin and may lead to the development of novel strategies for preventing dissemination of primary breast tumors predicted at the time of diagnosis to be highly metastatic, and of secondary tumors that have already spread to other parts of the body.
- Cell Migration and Invasion Laboratory, The Azrieli Faculty of Medicine, Bar-Ilan University, Safed, 1311502, Israel.
Organizational Affiliation: