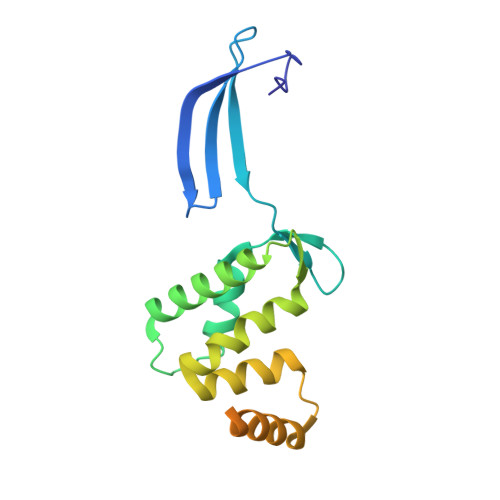
A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Rozman Grinberg, I., Martinez-Carranza, M., Bimai, O., Nouairia, G., Shahid, S., Lundin, D., Logan, D.T., Sjoberg, B.M., Stenmark, P.(2022) Nat Commun 13: 2700-2700
- PubMed: 35577776
- DOI: https://doi.org/10.1038/s41467-022-30328-1
- Primary Citation of Related Structures:
7P37, 7P3F, 7P3Q - PubMed Abstract:
Ribonucleotide reductase (RNR) is an essential enzyme that catalyzes the synthesis of DNA building blocks in virtually all living cells. NrdR, an RNR-specific repressor, controls the transcription of RNR genes and, often, its own, in most bacteria and some archaea. NrdR senses the concentration of nucleotides through its ATP-cone, an evolutionarily mobile domain that also regulates the enzymatic activity of many RNRs, while a Zn-ribbon domain mediates binding to NrdR boxes upstream of and overlapping the transcription start site of RNR genes. Here, we combine biochemical and cryo-EM studies of NrdR from Streptomyces coelicolor to show, at atomic resolution, how NrdR binds to DNA. The suggested mechanism involves an initial dodecamer loaded with two ATP molecules that cannot bind to DNA. When dATP concentrations increase, an octamer forms that is loaded with one molecule each of dATP and ATP per monomer. A tetramer derived from this octamer then binds to DNA and represses transcription of RNR. In many bacteria - including well-known pathogens such as Mycobacterium tuberculosis - NrdR simultaneously controls multiple RNRs and hence DNA synthesis, making it an excellent target for novel antibiotics development.
- Department of Biochemistry and Biophysics, Stockholm University, SE-10691, Stockholm, Sweden.
Organizational Affiliation: