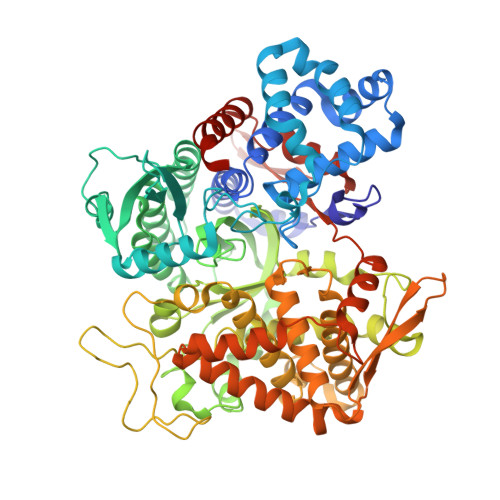
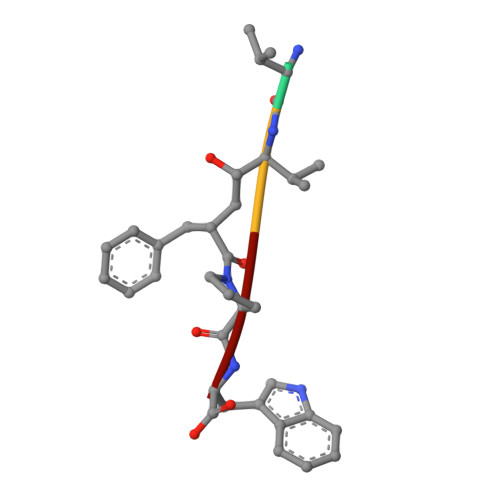
Efficient Entropy-Driven Inhibition of Dipeptidyl Peptidase III by Hydroxyethylene Transition-State Peptidomimetics.
Ivkovic, J., Jha, S., Lembacher-Fadum, C., Puschnig, J., Kumar, P., Reithofer, V., Gruber, K., Macheroux, P., Breinbauer, R.(2021) Chemistry 27: 14108-14120
- PubMed: 34314529
- DOI: https://doi.org/10.1002/chem.202102204
- Primary Citation of Related Structures:
7OUP - PubMed Abstract:
Dipeptidyl peptidase III (DPP3) is a ubiquitously expressed Zn-dependent protease, which plays an important role in regulating endogenous peptide hormones, such as enkephalins or angiotensins. In previous biophysical studies, it could be shown that substrate binding is driven by a large entropic contribution due to the release of water molecules from the closing binding cleft. Here, the design, synthesis and biophysical characterization of peptidomimetic inhibitors is reported, using for the first time an hydroxyethylene transition-state mimetic for a metalloprotease. Efficient routes for the synthesis of both stereoisomers of the pseudopeptide core were developed, which allowed the synthesis of peptidomimetic inhibitors mimicking the VVYPW-motif of tynorphin. The best inhibitors inhibit DPP3 in the low μM range. Biophysical characterization by means of ITC measurement and X-ray crystallography confirm the unusual entropy-driven mode of binding. Stability assays demonstrated the desired stability of these inhibitors, which efficiently inhibited DPP3 in mouse brain homogenate.
- Institute of Organic Chemistry, Graz University of Technology, Stremayrgasse 9, 8010, Graz, Austria.
Organizational Affiliation: