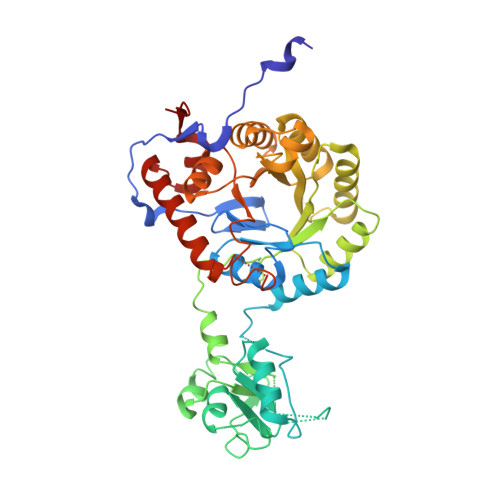
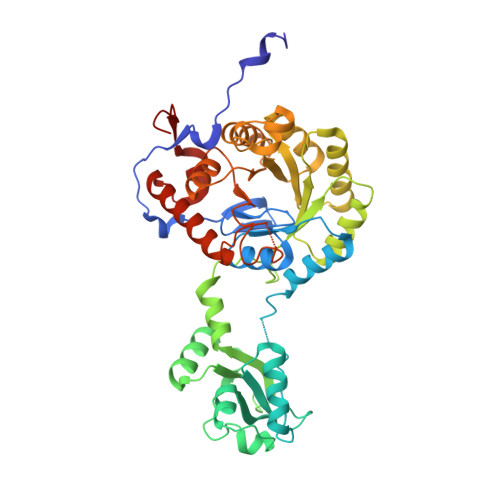
Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Giammarinaro, P.I., Young, M.K.M., Steinchen, W., Mais, C.N., Hochberg, G., Yang, J., Stevenson, D.M., Amador-Noguez, D., Paulus, A., Wang, J.D., Bange, G.(2022) Nat Microbiol 7: 1442-1452
- PubMed: 35953658
- DOI: https://doi.org/10.1038/s41564-022-01193-x
- Primary Citation of Related Structures:
7OJ1, 7OJ2 - PubMed Abstract:
Diadenosine tetraphosphate (Ap4A) is a putative second messenger molecule that is conserved from bacteria to humans. Nevertheless, its physiological role and the underlying molecular mechanisms are poorly characterized. We investigated the molecular mechanism by which Ap4A regulates inosine-5'-monophosphate dehydrogenase (IMPDH, a key branching point enzyme for the biosynthesis of adenosine or guanosine nucleotides) in Bacillus subtilis. We solved the crystal structure of BsIMPDH bound to Ap4A at a resolution of 2.45 Å to show that Ap4A binds to the interface between two IMPDH subunits, acting as the glue that switches active IMPDH tetramers into less active octamers. Guided by these insights, we engineered mutant strains of B. subtilis that bypass Ap4A-dependent IMPDH regulation without perturbing intracellular Ap4A pools themselves. We used metabolomics, which suggests that these mutants have a dysregulated purine, and in particular GTP, metabolome and phenotypic analysis, which shows increased sensitivity of B. subtilis IMPDH mutant strains to heat compared with wild-type strains. Our study identifies a central role for IMPDH in remodelling metabolism and heat resistance, and provides evidence that Ap4A can function as an alarmone.
- Department of Chemistry and Center for Synthetic Microbiology, Philipps University Marburg, Marburg, Germany.
Organizational Affiliation: