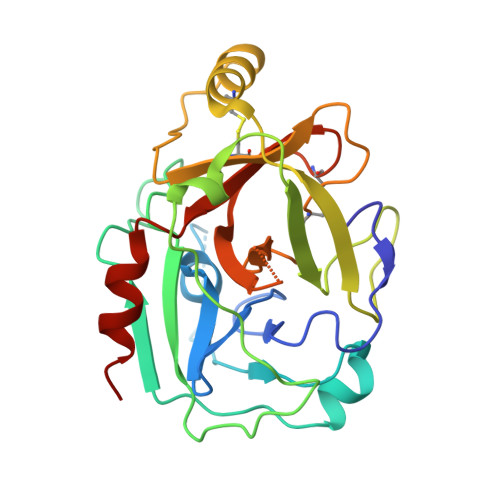
A Structural Basis for Inhibition of the Complement Initiator Protease C1r by Lyme Disease Spirochetes.
Garrigues, R.J., Powell-Pierce, A.D., Hammel, M., Skare, J.T., Garcia, B.L.(2021) J Immunol 207: 2856-2867
- PubMed: 34759015
- DOI: https://doi.org/10.4049/jimmunol.2100815
- Primary Citation of Related Structures:
7MZT - PubMed Abstract:
Complement evasion is a hallmark of extracellular microbial pathogens such as Borrelia burgdorferi , the causative agent of Lyme disease. Lyme disease spirochetes express nearly a dozen outer surface lipoproteins that bind complement components and interfere with their native activities. Among these, BBK32 is unique in its selective inhibition of the classical pathway. BBK32 blocks activation of this pathway by selectively binding and inhibiting the C1r serine protease of the first component of complement, C1. To understand the structural basis for BBK32-mediated C1r inhibition, we performed crystallography and size-exclusion chromatography-coupled small angle X-ray scattering experiments, which revealed a molecular model of BBK32-C in complex with activated human C1r. Structure-guided site-directed mutagenesis was combined with surface plasmon resonance binding experiments and assays of complement function to validate the predicted molecular interface. Analysis of the structures shows that BBK32 inhibits activated forms of C1r by occluding substrate interaction subsites (i.e., S1 and S1') and reveals a surprising role for C1r B loop-interacting residues for full inhibitory activity of BBK32. The studies reported in this article provide for the first time (to our knowledge) a structural basis for classical pathway-specific inhibition by a human pathogen.
- Department of Microbiology and Immunology, Brody School of Medicine, East Carolina University, Greenville, NC.
Organizational Affiliation: