Structure of an antennally-expressed carboxylesterase suggests lepidopteran odorant degrading enzymes are broadly tuned
Corcoran, J.A., Hamiaux, C., Faraone, N., Lofstedt, C., Carraher, C.(2023) Curr Res Insect Sci 3: 100062
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2023) Curr Res Insect Sci 3: 100062
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carboxylesterase-24 | 556 | Epiphyas postvittana | Mutation(s): 0 EC: 3.1.1.1 | 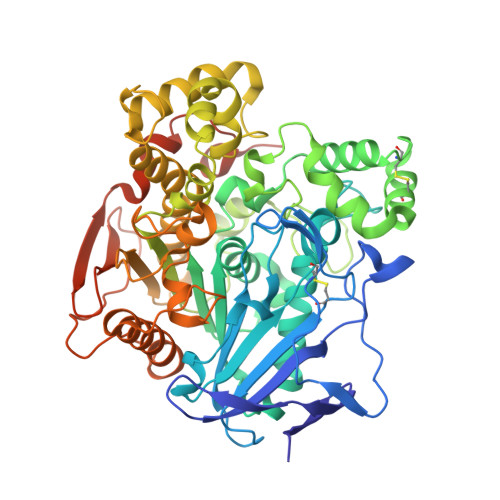 | |
UniProt | |||||
Find proteins for A0A0K8TUN4 (Epiphyas postvittana) Explore A0A0K8TUN4 Go to UniProtKB: A0A0K8TUN4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0K8TUN4 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | I, J, K, L, M I, J, K, L, M, N, O, P | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42227JK GlyCosmos: G42227JK GlyGen: G42227JK | |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 107.475 | α = 78.31 |
| b = 113.967 | β = 70.18 |
| c = 125.772 | γ = 65.88 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| Aimless | data scaling |
| MoRDa | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |