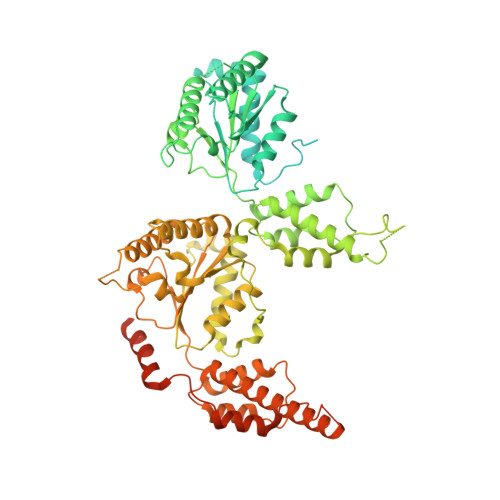
Active conformation of the p97-p47 unfoldase complex.
Xu, Y., Han, H., Cooney, I., Guo, Y., Moran, N.G., Zuniga, N.R., Price, J.C., Hill, C.P., Shen, P.S.(2022) Nat Commun 13: 2640-2640
- PubMed: 35552390
- DOI: https://doi.org/10.1038/s41467-022-30318-3
- Primary Citation of Related Structures:
7MHS - PubMed Abstract:
The p97 AAA+ATPase is an essential and abundant regulator of protein homeostasis that plays a central role in unfolding ubiquitylated substrates. Here we report two cryo-EM structures of human p97 in complex with its p47 adaptor. One of the conformations is six-fold symmetric, corresponds to previously reported structures of p97, and lacks bound substrate. The other structure adopts a helical conformation, displays substrate running in an extended conformation through the pore of the p97 hexamer, and resembles structures reported for other AAA unfoldases. These findings support the model that p97 utilizes a "hand-over-hand" mechanism in which two residues of the substrate are translocated for hydrolysis of two ATPs, one in each of the two p97 AAA ATPase rings. Proteomics analysis supports the model that one p97 complex can bind multiple substrate adaptors or binding partners, and can process substrates with multiple types of ubiquitin modification.
- Department of Biochemistry, 15 N. Medical Drive East, University of Utah, Salt Lake City, UT, 84112, USA.
Organizational Affiliation: