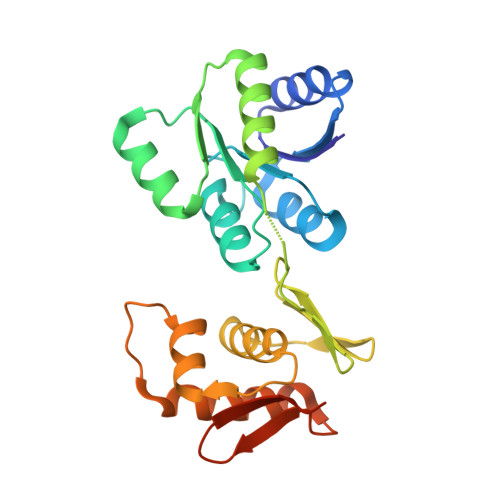
Structures of full-length VanR from Streptomyces coelicolor in both the inactive and activated states.
Maciunas, L.J., Porter, N., Lee, P.J., Gupta, K., Loll, P.J.(2021) Acta Crystallogr D Struct Biol 77: 1027-1039
- PubMed: 34342276
- DOI: https://doi.org/10.1107/S2059798321006288
- Primary Citation of Related Structures:
7LZ9, 7LZA - PubMed Abstract:
Vancomycin has historically been used as a last-resort treatment for serious bacterial infections. However, vancomycin resistance has become widespread in certain pathogens, presenting a serious threat to public health. Resistance to vancomycin is conferred by a suite of resistance genes, the expression of which is controlled by the VanR-VanS two-component system. VanR is the response regulator in this system; in the presence of vancomycin, VanR accepts a phosphoryl group from VanS, thereby activating VanR as a transcription factor and inducing expression of the resistance genes. This paper presents the X-ray crystal structures of full-length VanR from Streptomyces coelicolor in both the inactive and activated states at resolutions of 2.3 and 2.0 Å, respectively. Comparison of the two structures illustrates that phosphorylation of VanR is accompanied by a disorder-to-order transition of helix 4, which lies within the receiver domain of the protein. This transition generates an interface that promotes dimerization of the receiver domain; dimerization in solution was verified using analytical ultracentrifugation. The inactive conformation of the protein does not appear intrinsically unable to bind DNA; rather, it is proposed that in the activated form DNA binding is enhanced by an avidity effect contributed by the receiver-domain dimerization.
- Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, Philadelphia, PA 19102, USA.
Organizational Affiliation: