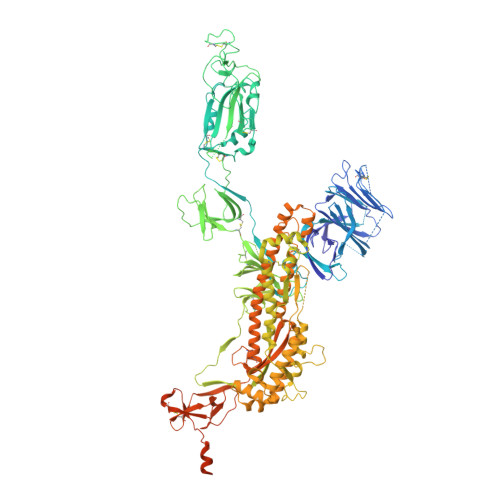
A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Xiao, T., Lu, J., Zhang, J., Johnson, R.I., McKay, L.G.A., Storm, N., Lavine, C.L., Peng, H., Cai, Y., Rits-Volloch, S., Lu, S., Quinlan, B.D., Farzan, M., Seaman, M.S., Griffiths, A., Chen, B.(2021) Nat Struct Mol Biol 28: 202-209
- PubMed: 33432247
- DOI: https://doi.org/10.1038/s41594-020-00549-3
- Primary Citation of Related Structures:
7KJ2, 7KJ3, 7KJ4, 7KJ5 - PubMed Abstract:
Effective intervention strategies are urgently needed to control the COVID-19 pandemic. Human angiotensin-converting enzyme 2 (ACE2) is a membrane-bound carboxypeptidase that forms a dimer and serves as the cellular receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). ACE2 is also a key negative regulator of the renin-angiotensin system that modulates vascular functions. We report here the properties of a trimeric ACE2 ectodomain variant, engineered using a structure-based approach. The trimeric ACE2 variant has a binding affinity of ~60 pM for the spike protein of SARS‑CoV‑2 (compared with 77 nM for monomeric ACE2 and 12-22 nM for dimeric ACE2 constructs), and its peptidase activity and the ability to block activation of angiotensin II receptor type 1 in the renin-angiotensin system are preserved. Moreover, the engineered ACE2 potently inhibits SARS‑CoV‑2 infection in cell culture. These results suggest that engineered, trimeric ACE2 may be a promising anti-SARS-CoV-2 agent for treating COVID-19.
- Division of Molecular Medicine, Boston Children's Hospital, Boston, MA, USA.
Organizational Affiliation: