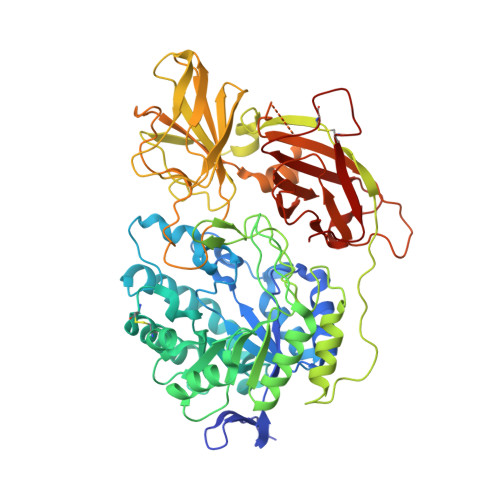
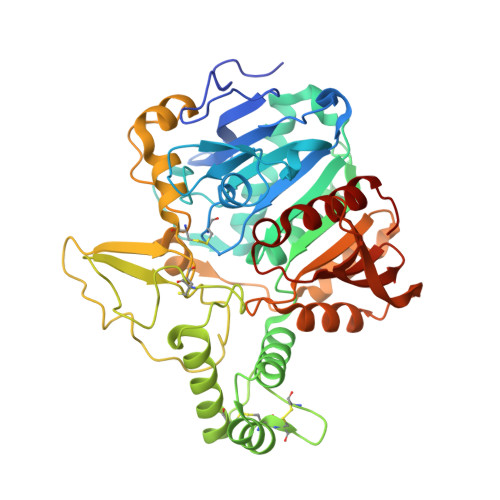
Structure of the murine lysosomal multienzyme complex core.
Gorelik, A., Illes, K., Hasan, S.M.N., Nagar, B., Mazhab-Jafari, M.T.(2021) Sci Adv 7
- PubMed: 33980489
- DOI: https://doi.org/10.1126/sciadv.abf4155
- Primary Citation of Related Structures:
7KDV - PubMed Abstract:
The enzymes β-galactosidase (GLB1) and neuraminidase 1 (NEU1; sialidase 1) participate in the degradation of glycoproteins and glycolipids in the lysosome. To remain active and stable, they associate with PPCA [protective protein cathepsin A (CTSA)] into a high-molecular weight lysosomal multienzyme complex (LMC), of which several forms exist. Genetic defects in these three proteins cause the lysosomal storage diseases GM1-gangliosidosis/mucopolysaccharidosis IV type B, sialidosis, and galactosialidosis, respectively. To better understand the interactions between these enzymes, we determined the three-dimensional structure of the murine LMC core. This 0.8-MDa complex is composed of three GLB1 dimers and three CTSA dimers, adopting a triangular architecture maintained through six copies of a unique GLB1-CTSA polar interface. Mutations in this contact surface that occur in GM1-gangliosidosis prevent formation of the LMC in vitro. These findings may facilitate development of therapies for lysosomal storage disorders.
- Department of Biochemistry, McGill University, Montreal, Quebec, Canada.
Organizational Affiliation: