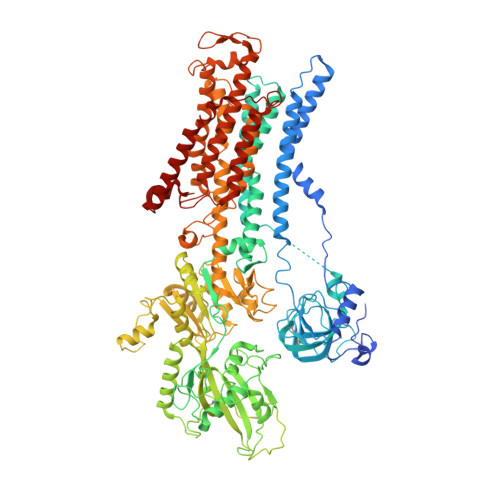
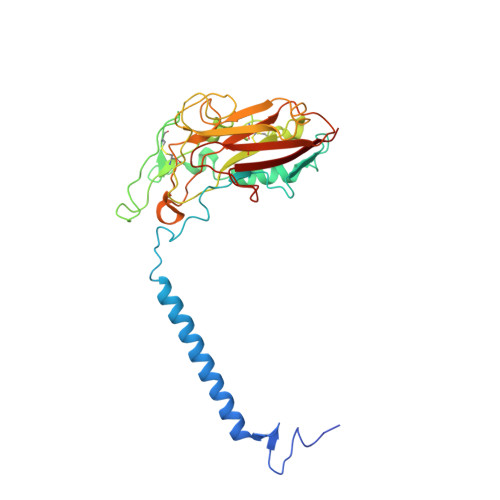
Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Guo, Y., Zhang, Y., Yan, R., Huang, B., Ye, F., Wu, L., Chi, X., Shi, Y., Zhou, Q.(2022) Nat Commun 13: 3957-3957
- PubMed: 35803952
- DOI: https://doi.org/10.1038/s41467-022-31602-y
- Primary Citation of Related Structures:
7E1Z, 7E20, 7E21 - PubMed Abstract:
Sodium-Potassium Pump (Na + /K + -ATPase, NKA) is an ion pump that generates an electrochemical gradient of sodium and potassium ions across the plasma membrane by hydrolyzing ATP. During each Post-Albers cycle, NKA exchanges three cytoplasmic sodium ions for two extracellular potassium ions through alternating changes between the E1 and E2 states. Hitherto, several steps remained unknown during the complete working cycle of NKA. Here, we report cryo-electron microscopy (cryo-EM) structures of recombinant human NKA (hNKA) in three distinct states at 2.7-3.2 Å resolution, representing the E1·3Na and E1·3Na·ATP states with cytosolic gates open and the basic E2·[2K] state, respectively. This work provides the insights into the cytoplasmic Na + entrance pathway and the mechanism of cytoplasmic gate closure coupled with ATP hydrolysis, filling crucial gaps in the structural elucidation of the Post-Albers cycle of NKA.
- Westlake Laboratory of Life Sciences and Biomedicine, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, 310024, Zhejiang Province, China.
Organizational Affiliation: