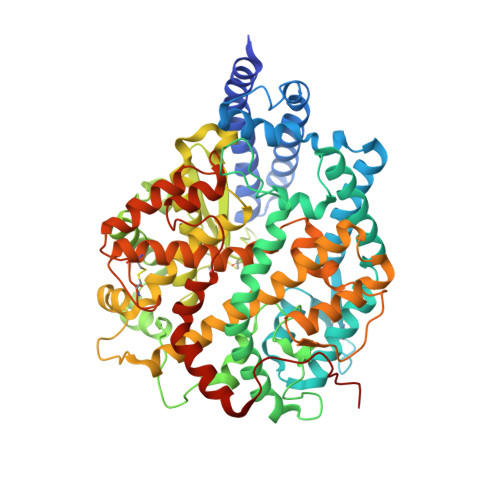
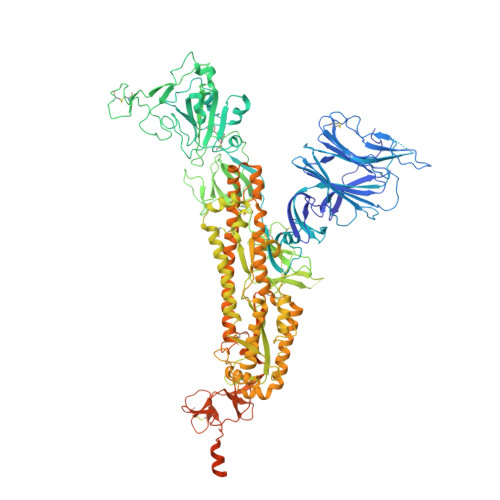
Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Xu, C., Wang, Y., Liu, C., Zhang, C., Han, W., Hong, X., Wang, Y., Hong, Q., Wang, S., Zhao, Q., Wang, Y., Yang, Y., Chen, K., Zheng, W., Kong, L., Wang, F., Zuo, Q., Huang, Z., Cong, Y.(2021) Sci Adv 7
- PubMed: 33277323
- DOI: https://doi.org/10.1126/sciadv.abe5575
- Primary Citation of Related Structures:
7DF3, 7DF4, 7DK3 - PubMed Abstract:
The recent outbreaks of SARS-CoV-2 pose a global health emergency. The SARS-CoV-2 trimeric spike (S) glycoprotein interacts with the human ACE2 receptor to mediate viral entry into host cells. We report the cryo-EM structures of a tightly closed SARS-CoV-2 S trimer with packed fusion peptide and an ACE2-bound S trimer at 2.7- and 3.8-Å resolution, respectively. Accompanying ACE2 binding to the up receptor-binding domain (RBD), the associated ACE2-RBD exhibits continuous swing motions. Notably, the SARS-CoV-2 S trimer appears much more sensitive to the ACE2 receptor than the SARS-CoV S trimer regarding receptor-triggered transformation from the closed prefusion state to the fusion-prone open state, potentially contributing to the superior infectivity of SARS-CoV-2. We defined the RBD T470-T478 loop and Y505 as viral determinants for specific recognition of SARS-CoV-2 RBD by ACE2. Our findings depict the mechanism of ACE2-induced S trimer conformational transitions from the ground prefusion state toward the postfusion state, facilitating development of anti-SARS-CoV-2 vaccines and therapeutics.
- State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: