Pentavalent Sialic Acid Conjugates Block Coxsackievirus A24 Variant and Human Adenovirus Type 37-Viruses That Cause Highly Contagious Eye Infections.
Johansson, E., Caraballo, R., Mistry, N., Zocher, G., Qian, W., Andersson, C.D., Hurdiss, D.L., Chandra, N., Thompson, R., Frangsmyr, L., Stehle, T., Arnberg, N., Elofsson, M.(2020) ACS Chem Biol 15: 2683-2691
- PubMed: 32845119
- DOI: https://doi.org/10.1021/acschembio.0c00446
- Primary Citation of Related Structures:
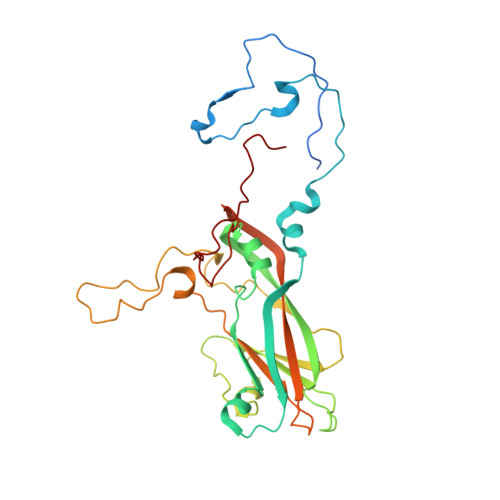
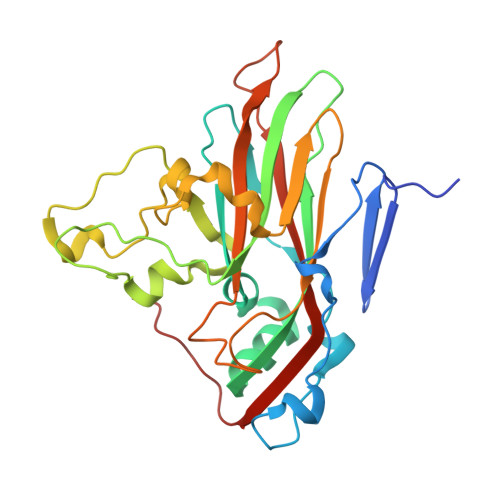
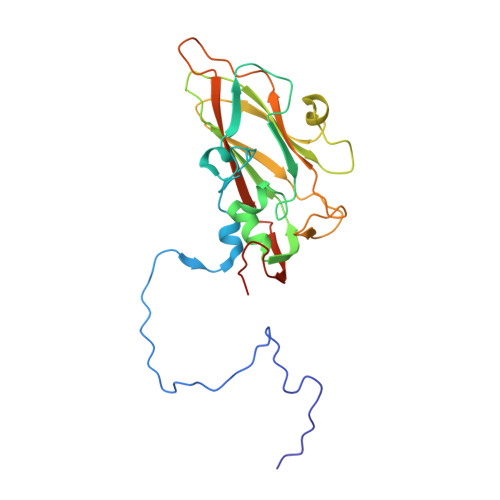
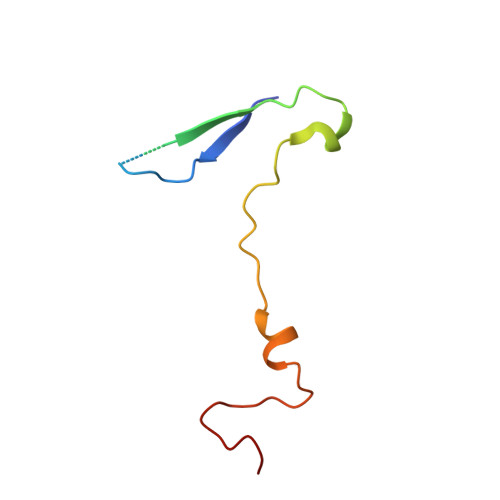
6TSD - PubMed Abstract:
Coxsackievirus A24 variant (CVA24v) and human adenovirus 37 (HAdV-37) are leading causative agents of the severe and highly contagious ocular infections acute hemorrhagic conjunctivitis and epidemic keratoconjunctivitis, respectively. Currently, neither vaccines nor antiviral agents are available for treating these diseases, which affect millions of individuals worldwide. CVA24v and HAdV-37 utilize sialic acid as attachment receptors facilitating entry into host cells. Previously, we and others have shown that derivatives based on sialic acid are effective in preventing HAdV-37 binding and infection of cells. Here, we designed and synthesized novel pentavalent sialic acid conjugates and studied their inhibitory effect against CVA24v and HAdV-37 binding and infection of human corneal epithelial cells. The pentavalent conjugates are the first reported inhibitors of CVA24v infection and proved efficient in blocking HAdV-37 binding. Taken together, the pentavalent conjugates presented here form a basis for the development of general inhibitors of these highly contagious ocular pathogens.
- Department of Chemistry, Umeå University, SE90187 Umeå, Sweden.
Organizational Affiliation: