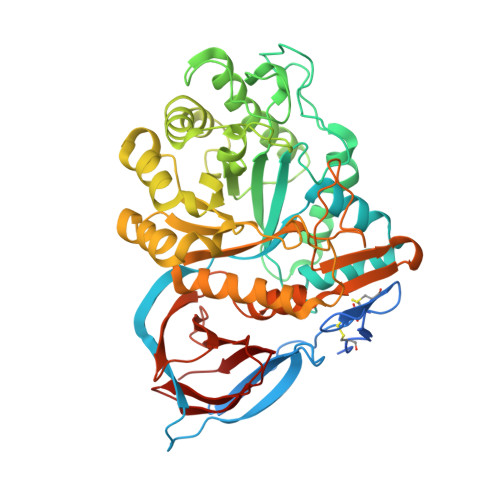
Novel beta-Glucocerebrosidase Activators That Bind to a New Pocket at a Dimer Interface and Induce Dimerization.
Benz, J., Rufer, A.C., Huber, S., Ehler, A., Hug, M., Topp, A., Guba, W., Hofmann, E.C., Jagasia, R., Rodriguez Sarmiento, R.M.(2021) Angew Chem Int Ed Engl 60: 5436-5442
- PubMed: 33238058
- DOI: https://doi.org/10.1002/anie.202013890
- Primary Citation of Related Structures:
6T13 - PubMed Abstract:
Genetic, preclinical and clinical data link Parkinson's disease and Gaucher's disease and provide a rational entry point to disease modification therapy via enhancement of β-Glucocerebrosidase (GCase) activity. We discovered a new class of pyrrolo[2,3-b]pyrazine activators effecting both Vmax and Km. They bind to human GCase and increase substrate metabolism in the lysosome in a cellular assay. We obtained the first crystal structure for an activator and identified a novel non-inhibitory binding mode at the interface of a dimer, rationalizing the observed structure-activity relationship (SAR). The compound binds GCase inducing formation of a dimeric state at both endoplasmic reticulum (ER) and lysosomal pHs, as confirmed by analytical ultracentrifugation. Importantly, the pyrrolo[2,3-b]pyrazines have central nervous system (CNS) drug-like properties. Our findings are important for future drug discovery efforts in the field of GCase activation and provide a deeper mechanistic understanding of the requirements for enzymatic activation, pointing to the relevance of dimerization.
- Lead Discovery, Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd., Grenzacherstrasse 124, 4070, Basel, Switzerland.
Organizational Affiliation: