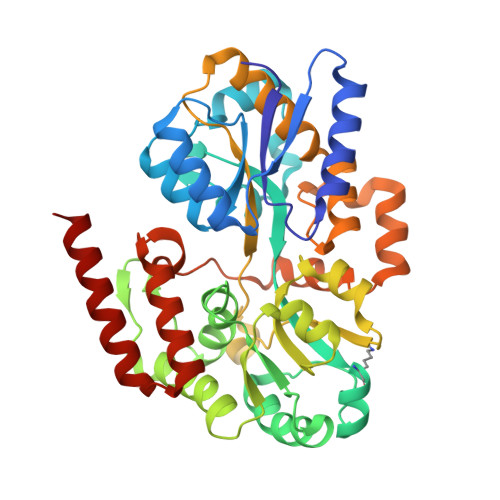
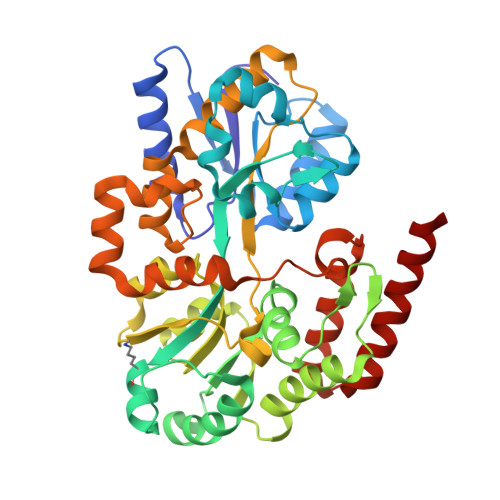
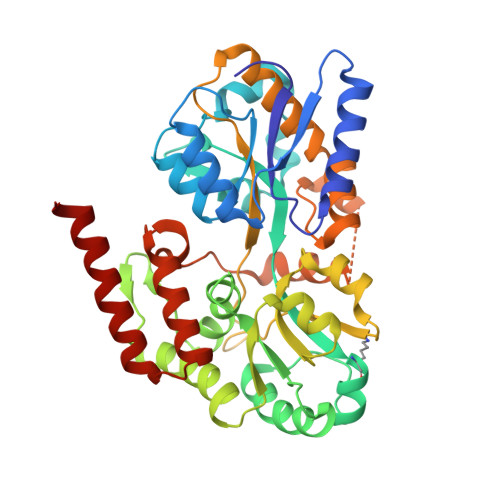
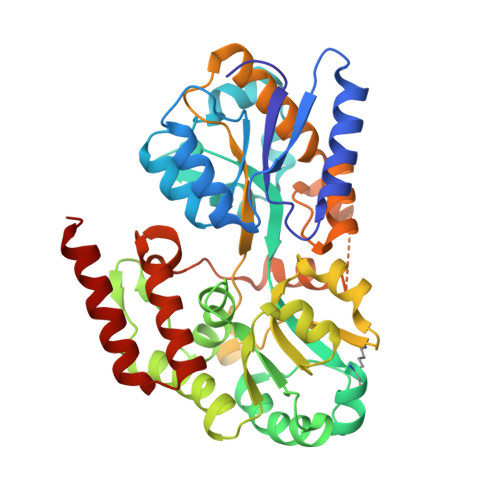
Structural Basis of Glycerophosphodiester Recognition by theMycobacterium tuberculosisSubstrate-Binding Protein UgpB.
Fenn, J.S., Nepravishta, R., Guy, C.S., Harrison, J., Angulo, J., Cameron, A.D., Fullam, E.(2019) ACS Chem Biol 14: 1879-1887
- PubMed: 31433162
- DOI: https://doi.org/10.1021/acschembio.9b00204
- Primary Citation of Related Structures:
6R1B - PubMed Abstract:
Mycobacterium tuberculosis ( Mtb ) is the causative agent of tuberculosis (TB) and has evolved an incredible ability to survive latently within the human host for decades. The Mtb pathogen encodes for a low number of ATP-binding cassette (ABC) importers for the acquisition of carbohydrates that may reflect the nutrient poor environment within the host macrophages. Mtb UgpB (Rv2833c) is the substrate binding domain of the UgpABCE transporter that recognizes glycerophosphocholine (GPC), indicating that this transporter has a role in recycling glycerophospholipid metabolites. By using a combination of saturation transfer difference (STD) NMR and X-ray crystallography, we report the structural analysis of Mtb UgpB complexed with GPC and have identified that Mtb UgpB not only recognizes GPC but is also promiscuous for a broad range of glycerophosphodiesters. Complementary biochemical analyses and site-directed mutagenesis precisely define the molecular basis and specificity of glycerophosphodiester recognition. Our results provide critical insights into the structural and functional role of the Mtb UgpB transporter and reveal that the specificity of this ABC-transporter is not limited to GPC, therefore optimizing the ability of Mtb to scavenge scarce nutrients and essential glycerophospholipid metabolites via a single transporter during intracellular infection.
- School of Life Sciences , University of Warwick , Coventry , West Midlands CV4 7AL , United Kingdom.
Organizational Affiliation: