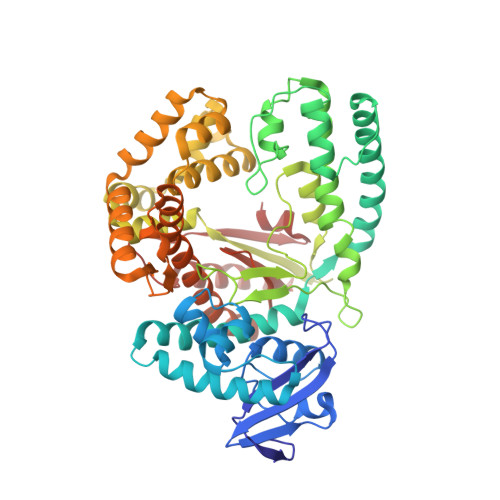
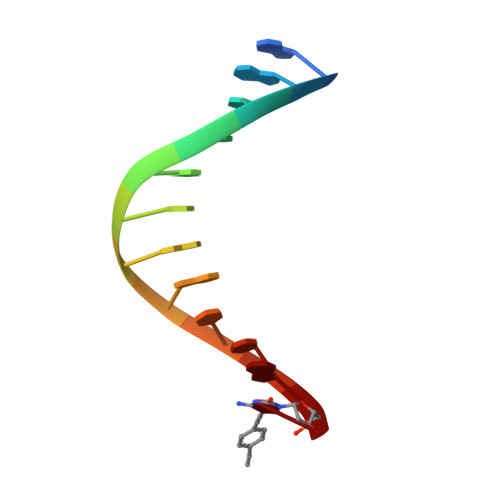
Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Kropp, H.M., Durr, S.L., Peter, C., Diederichs, K., Marx, A.(2018) Proc Natl Acad Sci U S A 115: 9992-9997
- PubMed: 30224478
- DOI: https://doi.org/10.1073/pnas.1811518115
- Primary Citation of Related Structures:
6FBC, 6FBD, 6FBE, 6FBF, 6FBG, 6FBH, 6FBI - PubMed Abstract:
DNA polymerases have evolved to process the four canonical nucleotides accurately. Nevertheless, these enzymes are also known to process modified nucleotides, which is the key to numerous core biotechnology applications. Processing of modified nucleotides includes incorporation of the modified nucleotide and postincorporation elongation to proceed with the synthesis of the nascent DNA strand. The structural basis for postincorporation elongation is currently unknown. We addressed this issue and successfully crystallized KlenTaq DNA polymerase in six closed ternary complexes containing the enzyme, the modified DNA substrate, and the incoming nucleotide. Each structure shows a high-resolution snapshot of the elongation of a modified primer, where the modification "moves" from the 3'-primer terminus upstream to the sixth nucleotide in the primer strand. Combining these data with quantum mechanics/molecular mechanics calculations and biochemical studies elucidates how the enzyme and the modified substrate mutually modulate their conformations without compromising the enzyme's activity significantly. The study highlights the plasticity of the system as origin of the broad substrate properties of DNA polymerases and facilitates the design of improved systems.
- Department of Chemistry, Universität Konstanz, 78464 Konstanz, Germany.
Organizational Affiliation: