Engineering Improved Antiphosphotyrosine Antibodies Based on an Immunoconvergent Binding Motif.
Mou, Y., Zhou, X.X., Leung, K., Martinko, A.J., Yu, J.Y., Chen, W., Wells, J.A.(2018) J Am Chem Soc 140: 16615-16624
- PubMed: 30398859
- DOI: https://doi.org/10.1021/jacs.8b08402
- Primary Citation of Related Structures:
6DEZ, 6DF0, 6DF1, 6DF2 - PubMed Abstract:
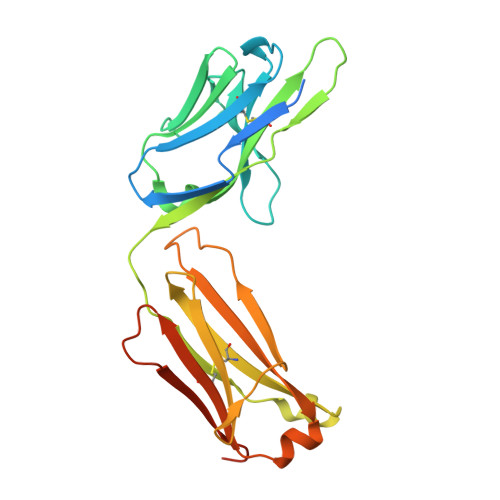
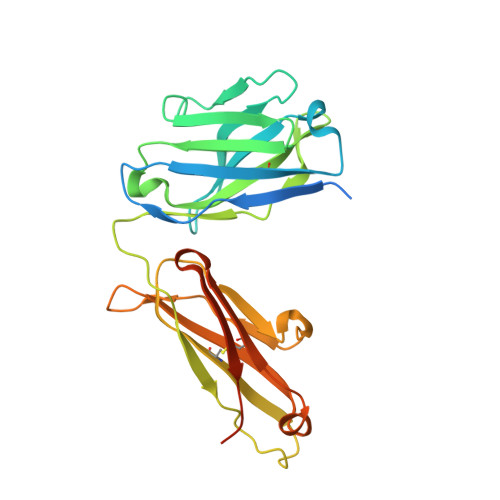
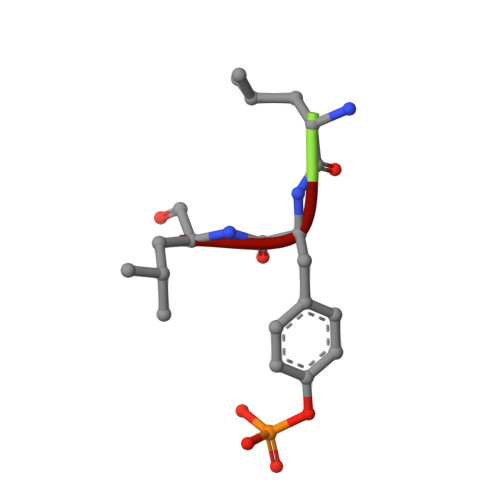
Phosphotyrosine (pY) is one of the most highly studied posttranslational modifications that is responsible for tightly regulating many signaling pathways in eukaryotes. Pan-specific pY antibodies have emerged as powerful tools for understanding the role of these modifications. Nevertheless, structures have not been reported for pan-specific pY antibodies, greatly impeding the further development of tools for integrating this ubiquitous posttranslational modification using structure-guided designs. Here, we present the first crystal structures of two widely utilized pan-specific pY antibodies, PY20 and 4G10. The two antibodies, although developed independently from animal immunizations, have surprisingly similar modes of recognition of the phosphate group, implicating a generic binding structure among pan-specific pY antibodies. Sequence alignments revealed that many pY binding residues are predominant in the mouse V germline genes, which consequently led to the convergent antibodies. On the basis of the convergent structure, we designed a phage display library by lengthening the CDR-L3 loop with the aid of computational modeling. Panning with this library resulted in a series of 4G10 variants with 4 to 11-fold improvements in pY binding affinities. The crystal structure of one improved variant showed remarkable superposition to the computational model, where the lengthened CDR-L3 loop creates an additional hydrogen bond indirectly bound to the phosphate group via a water molecule. The engineered variants exhibited superior performance in Western blot and immunofluorescence.
- Department of Pharmaceutical Chemistry , University of California, San Francisco , San Francisco , California 94143 , United States.
Organizational Affiliation: