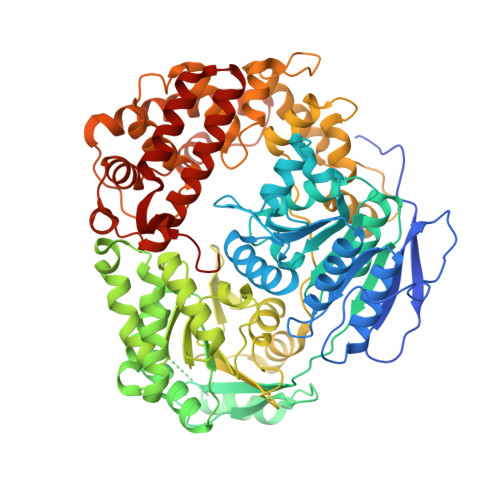
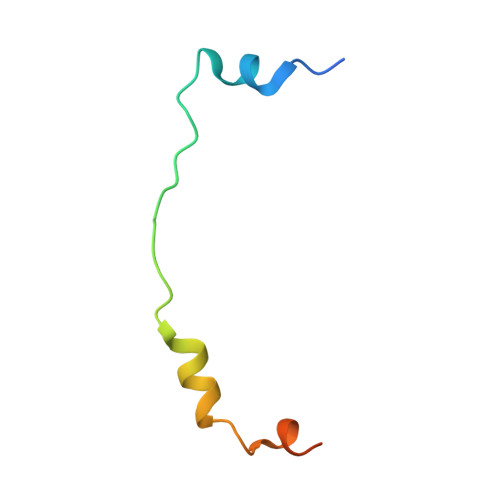
Structural basis for MTR4-ZCCHC8 interactions that stimulate the MTR4 helicase in the nuclear exosome-targeting complex.
Puno, M.R., Lima, C.D.(2018) Proc Natl Acad Sci U S A 115: E5506-E5515
- PubMed: 29844170
- DOI: https://doi.org/10.1073/pnas.1803530115
- Primary Citation of Related Structures:
6C90 - PubMed Abstract:
The nuclear exosome-targeting (NEXT) complex functions as an RNA exosome cofactor and is involved in surveillance and turnover of aberrant transcripts and noncoding RNAs. NEXT is a ternary complex composed of the RNA-binding protein RBM7, the scaffold zinc-knuckle protein ZCCHC8, and the helicase MTR4. While RNA interactions with RBM7 are known, it remains unclear how NEXT subunits collaborate to recognize and prepare substrates for degradation. Here, we show that MTR4 helicase activity is enhanced when associated with RBM7 and ZCCHC8. While uridine-rich substrates interact with RBM7 and are preferred, optimal activity is observed when substrates include a polyadenylated 3' end. We identify a bipartite interaction of ZCCHC8 with MTR4 and uncover a role for the conserved C-terminal domain of ZCCHC8 in stimulating MTR4 helicase and ATPase activities. A crystal structure reveals that the ZCCHC8 C-terminal domain binds the helicase core in a manner that is distinct from that observed for Saccharomyces cerevisiae exosome cofactors Trf4p and Air2p. Our results are consistent with a model whereby effective targeting of substrates by NEXT entails recognition of elements within the substrate and activation of MTR4 helicase activity.
- Structural Biology Program, Sloan Kettering Institute, New York, NY 10065.
Organizational Affiliation: