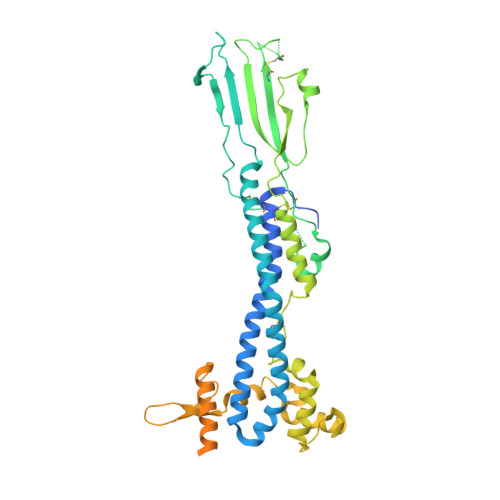
Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Zeelen, J., van Straaten, M., Verdi, J., Hempelmann, A., Hashemi, H., Perez, K., Jeffrey, P.D., Halg, S., Wiedemar, N., Maser, P., Papavasiliou, F.N., Stebbins, C.E.(2021) Nat Microbiol 6: 392-400
- PubMed: 33462435
- DOI: https://doi.org/10.1038/s41564-020-00844-1
- Primary Citation of Related Structures:
6Z79, 6Z7B, 6Z7C, 6Z7D, 6Z7E, 6Z8G, 6Z8H - PubMed Abstract:
Suramin has been a primary early-stage treatment for African trypanosomiasis for nearly 100 yr. Recent studies revealed that trypanosome strains that express the variant surface glycoprotein (VSG) VSGsur possess heightened resistance to suramin. Here, we show that VSGsur binds tightly to suramin but other VSGs do not. By solving high-resolution crystal structures of VSGsur and VSG13, we also demonstrate that these VSGs define a structurally divergent subgroup of the coat proteins. The co-crystal structure of VSGsur with suramin reveals that the chemically symmetric drug binds within a large cavity in the VSG homodimer asymmetrically, primarily through contacts of its central benzene rings. Structure-based, loss-of-contact mutations in VSGsur significantly decrease the affinity to suramin and lead to a loss of the resistance phenotype. Altogether, these data show that the resistance phenotype is dependent on the binding of suramin to VSGsur, establishing that the VSG proteins can possess functionality beyond their role in antigenic variation.
- Division of Structural Biology of Infection and Immunity, German Cancer Research Center, Heidelberg, Germany.
Organizational Affiliation: