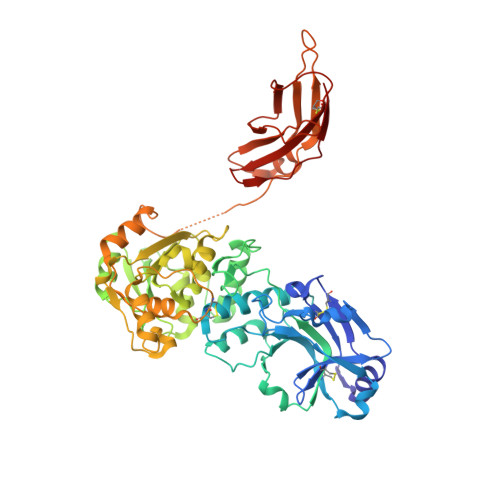
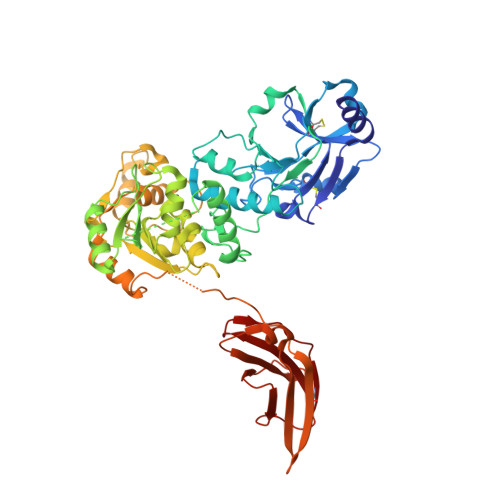
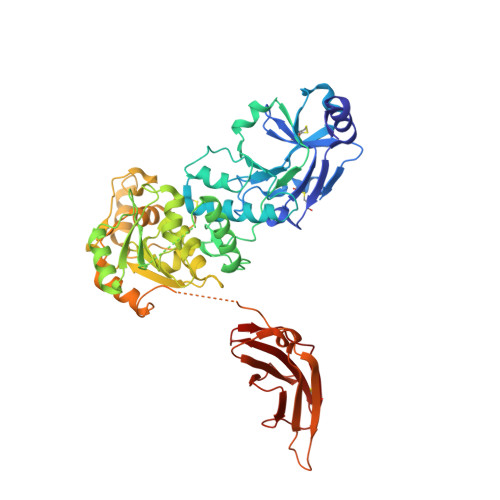
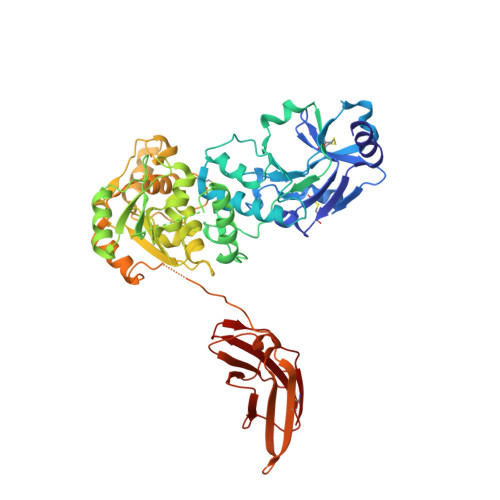
Crystal structures of beta-1,4-N-acetylglucosaminyltransferase 2: structural basis for inherited muscular dystrophies.
Yang, J.Y., Halmo, S.M., Praissman, J., Chapla, D., Singh, D., Wells, L., Moremen, K.W., Lanzilotta, W.N.(2021) Acta Crystallogr D Struct Biol 77: 486-495
- PubMed: 33825709
- DOI: https://doi.org/10.1107/S2059798321001261
- Primary Citation of Related Structures:
6XFI, 6XI2 - PubMed Abstract:
The canonical O-mannosylation pathway in humans is essential for the functional glycosylation of α-dystroglycan. Disruption of this post-translational modification pathway leads to congenital muscular dystrophies. The first committed step in the construction of a functional matriglycan structure involves the post-translational modification of α-dystroglycan. This is essential for binding extracellular matrix proteins and arenaviruses, and is catalyzed by β-1,4-N-acetylglucosaminyltransferase 2 (POMGNT2). While another glycosyl transferase, β-1,4-N-acetylglucosaminyltransferase 1 (POMGNT1), has been shown to be promiscuous in extending O-mannosylated sites, POMGNT2 has been shown to display significant primary amino-acid selectivity near the site of O-mannosylation. Moreover, several single point mutations in POMGNT2 have been identified in patients with assorted dystroglycanopathies such as Walker-Warburg syndrome and limb girdle muscular dystrophy. To gain insight into POMGNT2 function in humans, the enzyme was expressed as a soluble, secreted fusion protein by transient infection of HEK293 suspension cultures. Here, crystal structures of POMGNT2 (amino-acid residues 25-580) with and without UDP bound are reported. Consistent with a novel fold and a unique domain organization, no molecular-replacement model was available and phases were obtained through crystallization of a selenomethionine variant of the enzyme in the same space group. Tetragonal (space group P42 1 2; unit-cell parameters a = b = 129.8, c = 81.6 Å, α = γ = β = 90°) crystals with UDP bound diffracted to 1.98 Å resolution and contained a single monomer in the asymmetric unit. Orthorhombic (space group P2 1 2 1 2 1 ; unit-cell parameters a = 142.3, b = 153.9, c = 187.4 Å, α = γ = β = 90°) crystals were also obtained; they diffracted to 2.57 Å resolution and contained four monomers with differential glycosylation patterns and conformations. These structures provide the first rational basis for an explanation of the loss-of-function mutations and offer significant insights into the mechanics of this important human enzyme.
- The Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA.
Organizational Affiliation: