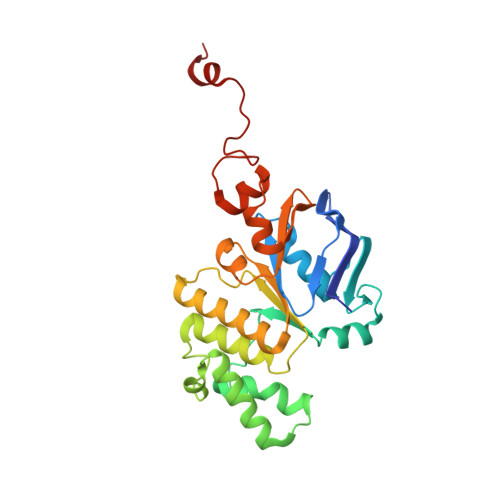
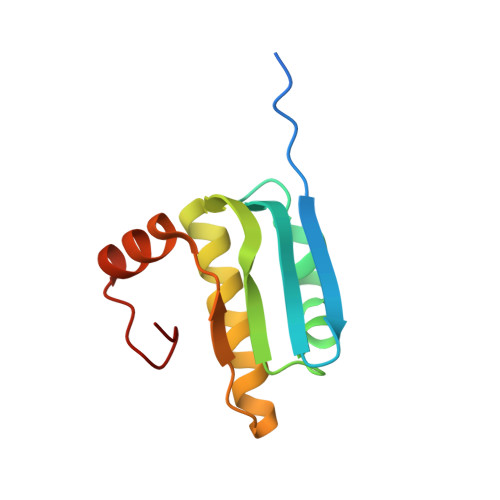
Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Kolich, L.R., Chang, Y.T., Coudray, N., Giacometti, S.I., MacRae, M.R., Isom, G.L., Teran, E.M., Bhabha, G., Ekiert, D.C.(2020) Elife 9
- PubMed: 32602838
- DOI: https://doi.org/10.7554/eLife.60030
- Primary Citation of Related Structures:
6XGY, 6XGZ - PubMed Abstract:
ABC transporters facilitate the movement of diverse molecules across cellular membranes, but how their activity is regulated post-translationally is not well understood. Here we report the crystal structure of MlaFB from E. coli , the cytoplasmic portion of the larger MlaFEDB ABC transporter complex, which drives phospholipid trafficking across the bacterial envelope to maintain outer membrane integrity. MlaB, a STAS domain protein, binds the ABC nucleotide binding domain, MlaF, and is required for its stability. Our structure also implicates a unique C-terminal tail of MlaF in self-dimerization. Both the C-terminal tail of MlaF and the interaction with MlaB are required for the proper assembly of the MlaFEDB complex and its function in cells. This work leads to a new model for how an important bacterial lipid transporter may be regulated by small proteins, and raises the possibility that similar regulatory mechanisms may exist more broadly across the ABC transporter family.
- Department of Cell Biology, New York University School of Medicine, New York, United States.
Organizational Affiliation: