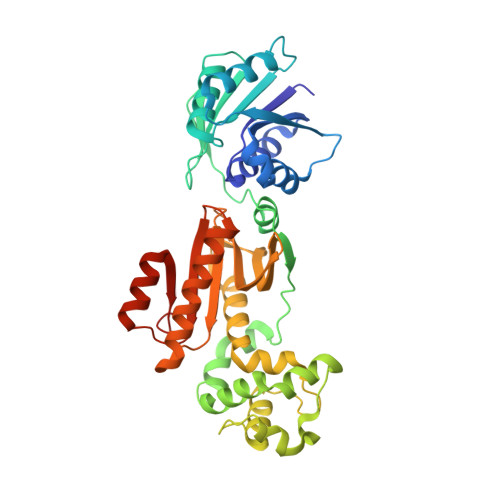
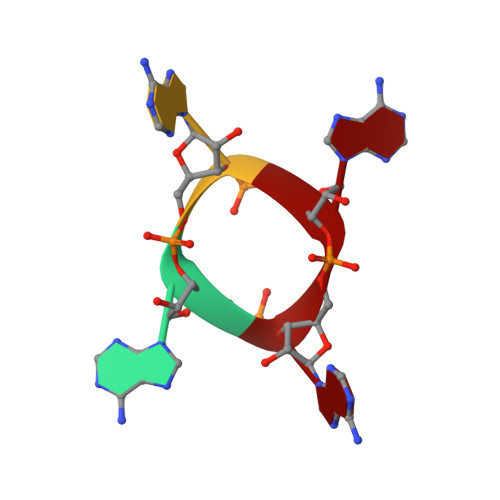
The Card1 nuclease provides defence during type III CRISPR immunity.
Rostol, J.T., Xie, W., Kuryavyi, V., Maguin, P., Kao, K., Froom, R., Patel, D.J., Marraffini, L.A.(2021) Nature 590: 624-629
- PubMed: 33461211
- DOI: https://doi.org/10.1038/s41586-021-03206-x
- Primary Citation of Related Structures:
6WXW, 6WXX, 6WXY, 6XL1 - PubMed Abstract:
In the type III CRISPR-Cas immune response of prokaryotes, infection triggers the production of cyclic oligoadenylates that bind and activate proteins that contain a CARF domain 1,2 . Many type III loci are associated with proteins in which the CRISPR-associated Rossman fold (CARF) domain is fused to a restriction endonuclease-like domain 3,4 . However, with the exception of the well-characterized Csm6 and Csx1 ribonucleases 5,6 , whether and how these inducible effectors provide defence is not known. Here we investigated a type III CRISPR accessory protein, which we name cyclic-oligoadenylate-activated single-stranded ribonuclease and single-stranded deoxyribonuclease 1 (Card1). Card1 forms a symmetrical dimer that has a large central cavity between its CRISPR-associated Rossmann fold and restriction endonuclease domains that binds cyclic tetra-adenylate. The binding of ligand results in a conformational change comprising the rotation of individual monomers relative to each other to form a more compact dimeric scaffold, in which a manganese cation coordinates the catalytic residues and activates the cleavage of single-stranded-but not double-stranded-nucleic acids (both DNA and RNA). In vivo, activation of Card1 induces dormancy of the infected hosts to provide immunity against phage infection and plasmids. Our results highlight the diversity of strategies used in CRISPR systems to provide immunity.
- Laboratory of Bacteriology, The Rockefeller University, New York, NY, USA.
Organizational Affiliation: