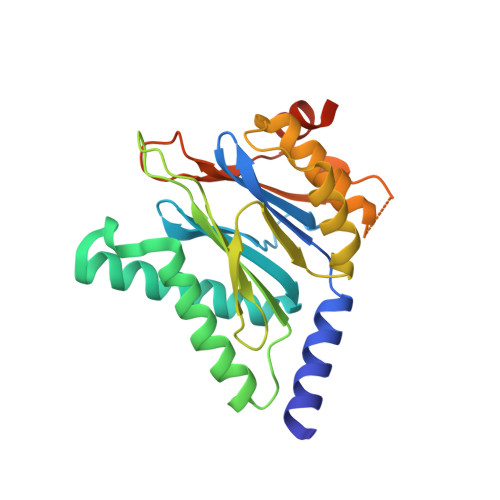
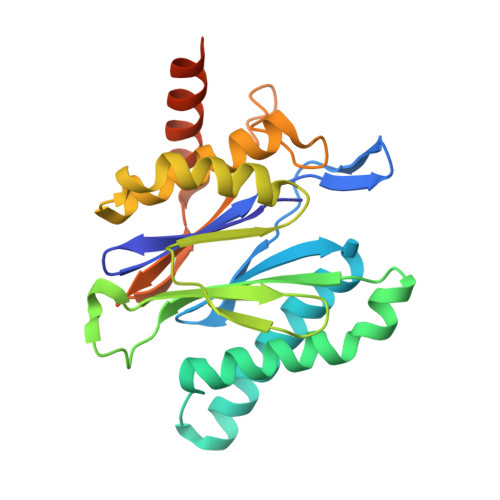
Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
Zhang, H., Hsu, H.C., Kahne, S.C., Hara, R., Zhan, W., Jiang, X., Burns-Huang, K., Ouellette, T., Imaeda, T., Okamoto, R., Kawasaki, M., Michino, M., Wong, T.T., Toita, A., Yukawa, T., Moraca, F., Vendome, J., Saha, P., Sato, K., Aso, K., Ginn, J., Meinke, P.T., Foley, M., Nathan, C.F., Darwin, K.H., Li, H., Lin, G.(2021) J Med Chem 64: 6262-6272
- PubMed: 33949190
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00296
- Primary Citation of Related Structures:
6WNK - PubMed Abstract:
Treatment of tuberculosis (TB) currently takes at least 6 months. Latent Mycobacterium tuberculosis (Mtb) is phenotypically tolerant to most anti-TB drugs. A key hypothesis is that drugs that kill nonreplicating (NR) Mtb may shorten treatment when used in combination with conventional drugs. The Mtb proteasome (Mtb20S) could be such a target because its pharmacological inhibition kills NR Mtb and its genetic deletion renders Mtb unable to persist in mice. Here, we report a series of macrocyclic peptides that potently and selectively target the Mtb20S over human proteasomes, including macrocycle 6 . The cocrystal structure of macrocycle 6 with Mtb20S revealed structural bases for the species selectivity. Inhibition of 20S within Mtb by 6 dose dependently led to the accumulation of Pup-tagged GFP that is degradable but resistant to depupylation and death of nonreplicating Mtb under nitrosative stress. These results suggest that compounds of this class have the potential to develop as anti-TB therapeutics.
- Department of Microbiology & Immunology, Weill Cornell Medicine, 1300 York Avenue, New York, New York 10065, United States.
Organizational Affiliation: