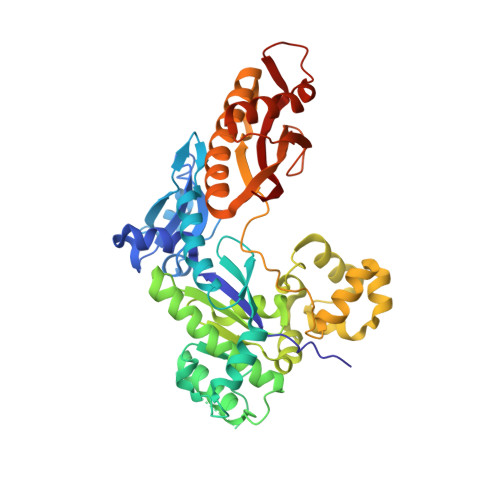
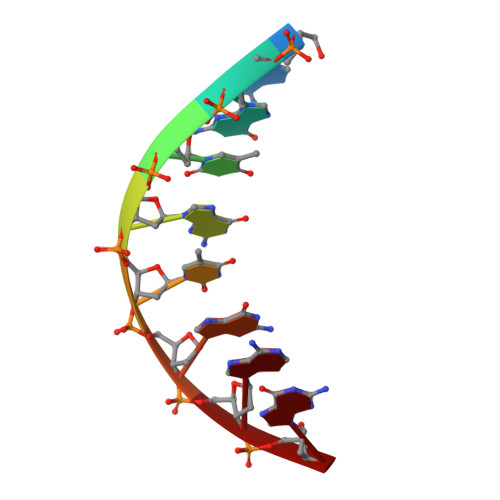
Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Jung, H., Hawkins, M., Lee, S.(2020) Biochem J 477: 4797-4810
- PubMed: 33258913
- DOI: https://doi.org/10.1042/BCJ20200800
- Primary Citation of Related Structures:
6MQ8, 6WK6 - PubMed Abstract:
The exocyclic amines of nucleobases can undergo deamination by various DNA damaging agents such as reactive oxygen species, nitric oxide, and water. The deamination of guanine and adenine generates the promutagenic xanthine and hypoxanthine, respectively. The exocyclic amines of bases in DNA are hydrogen bond donors, while the carbonyl moiety generated by the base deamination acts as hydrogen bond acceptors, which can alter base pairing properties of the purines. Xanthine is known to base pair with both cytosine and thymine, while hypoxanthine predominantly pairs with cytosine to promote A to G mutations. Despite the known promutagenicity of the major deaminated purines, structures of DNA polymerase bypassing these lesions have not been reported. To gain insights into the deaminated-induced mutagenesis, we solved crystal structures of human DNA polymerase η (polη) catalyzing across xanthine and hypoxanthine. In the catalytic site of polη, the deaminated guanine (i.e., xanthine) forms three Watson-Crick-like hydrogen bonds with an incoming dCTP, indicating the O2-enol tautomer of xanthine involves in the base pairing. The formation of the enol tautomer appears to be promoted by the minor groove contact by Gln38 of polη. When hypoxanthine is at the templating position, the deaminated adenine uses its O6-keto tautomer to form two Watson-Crick hydrogen bonds with an incoming dCTP, providing the structural basis for the high promutagenicity of hypoxanthine.
- The University of Texas at Austin, Austin, Texas, United States.
Organizational Affiliation: