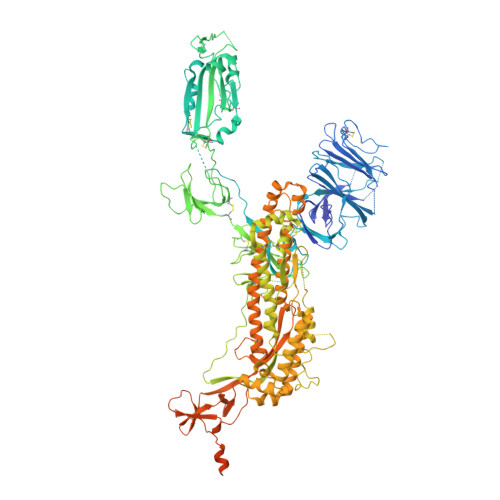
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.
Wrapp, D., Wang, N., Corbett, K.S., Goldsmith, J.A., Hsieh, C.L., Abiona, O., Graham, B.S., McLellan, J.S.(2020) Science 367: 1260-1263
- PubMed: 32075877
- DOI: https://doi.org/10.1126/science.abb2507
- Primary Citation of Related Structures:
6VSB - PubMed Abstract:
The outbreak of a novel coronavirus (2019-nCoV) represents a pandemic threat that has been declared a public health emergency of international concern. The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics. To facilitate medical countermeasure development, we determined a 3.5-angstrom-resolution cryo-electron microscopy structure of the 2019-nCoV S trimer in the prefusion conformation. The predominant state of the trimer has one of the three receptor-binding domains (RBDs) rotated up in a receptor-accessible conformation. We also provide biophysical and structural evidence that the 2019-nCoV S protein binds angiotensin-converting enzyme 2 (ACE2) with higher affinity than does severe acute respiratory syndrome (SARS)-CoV S. Additionally, we tested several published SARS-CoV RBD-specific monoclonal antibodies and found that they do not have appreciable binding to 2019-nCoV S, suggesting that antibody cross-reactivity may be limited between the two RBDs. The structure of 2019-nCoV S should enable the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: