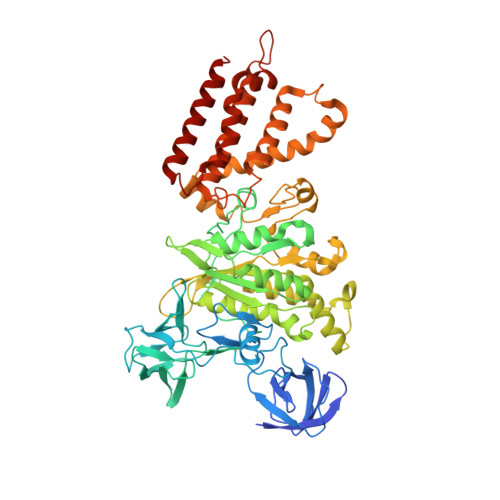
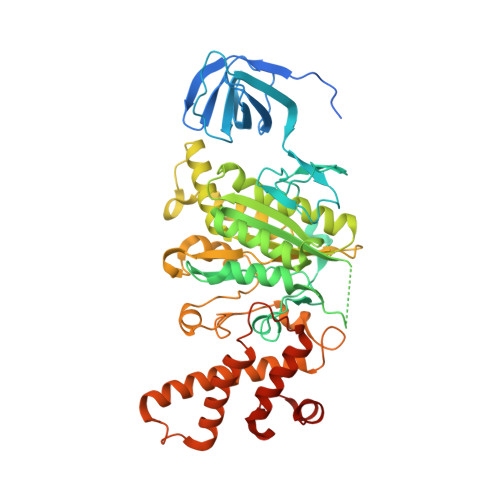
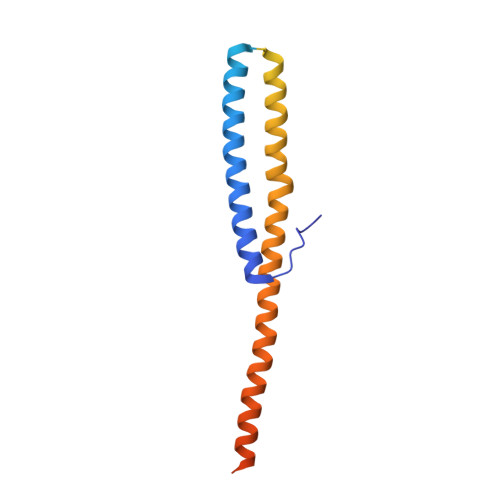
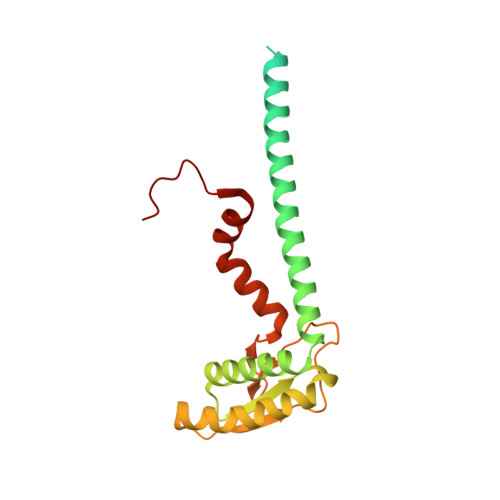
Structure of V-ATPase from the mammalian brain.
Abbas, Y.M., Wu, D., Bueler, S.A., Robinson, C.V., Rubinstein, J.L.(2020) Science 367: 1240-1246
- PubMed: 32165585
- DOI: https://doi.org/10.1126/science.aaz2924
- Primary Citation of Related Structures:
6VQ6, 6VQ7, 6VQ8, 6VQ9, 6VQA, 6VQB, 6VQC, 6VQG, 6VQH, 6VQI, 6VQJ, 6VQK - PubMed Abstract:
In neurons, the loading of neurotransmitters into synaptic vesicles uses energy from proton-pumping vesicular- or vacuolar-type adenosine triphosphatases (V-ATPases). These membrane protein complexes possess numerous subunit isoforms, which complicates their analysis. We isolated homogeneous rat brain V-ATPase through its interaction with SidK, a Legionella pneumophila effector protein. Cryo-electron microscopy allowed the construction of an atomic model, defining the enzyme's ATP:proton ratio as 3:10 and revealing a homolog of yeast subunit f in the membrane region, which we tentatively identify as RNAseK. The c ring encloses the transmembrane anchors for cleaved ATP6AP1/Ac45 and ATP6AP2/PRR, the latter of which is the (pro)renin receptor that, in other contexts, is involved in both Wnt signaling and the renin-angiotensin system that regulates blood pressure. This structure shows how ATP6AP1/Ac45 and ATP6AP2/PRR enable assembly of the enzyme's catalytic and membrane regions.
- Molecular Medicine Program, The Hospital for Sick Children Research Institute, Toronto, ON M5G 0A4, Canada.
Organizational Affiliation: