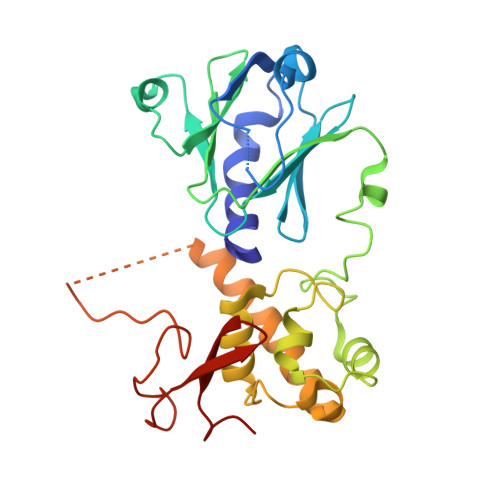
Conformational changes of DNA repair glycosylase MutM triggered by DNA binding.
Landova, B., Silhan, J.(2020) FEBS Lett 594: 3032-3044
- PubMed: 32598485
- DOI: https://doi.org/10.1002/1873-3468.13876
- Primary Citation of Related Structures:
6TC6, 6TC9 - PubMed Abstract:
Bacterial MutM is a DNA repair glycosylase removing DNA damage generated from oxidative stress and, therefore, preventing mutations and genomic instability. MutM belongs to the Fpg/Nei family of prokaryotic enzymes sharing structural and functional similarities with their eukaryotic counterparts, for example, NEIL1-NEIL3. Here, we present two crystal structures of MutM from pathogenic Neisseria meningitidis: a MutM holoenzyme and MutM bound to DNA. The free enzyme exists in an open conformation, while upon binding to DNA, both the enzyme and DNA undergo substantial structural changes and domain rearrangement. Our data show that not only NEI glycosylases but also the MutMs undergo dramatic conformational changes. Moreover, crystallographic data support the previously published observations that MutM enzymes are rather flexible and dynamic molecules.
- Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague, Czech Republic.
Organizational Affiliation: