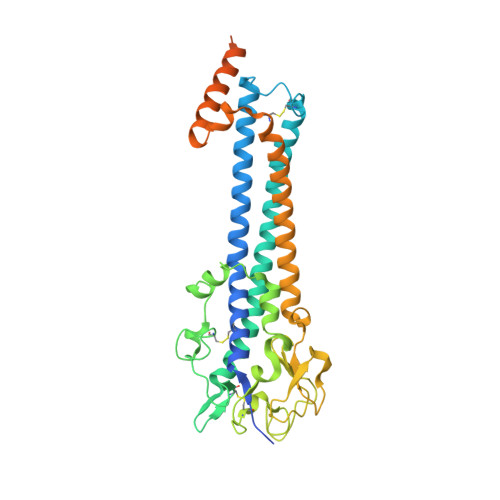
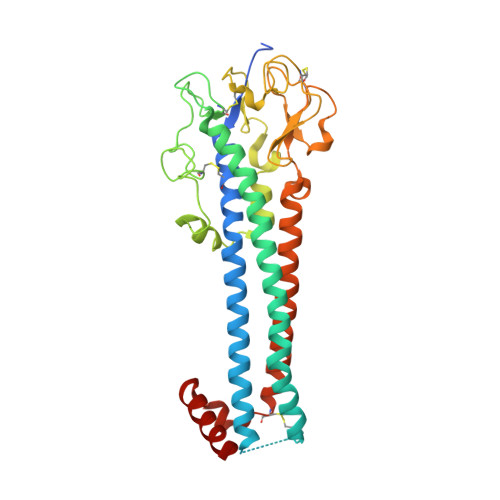
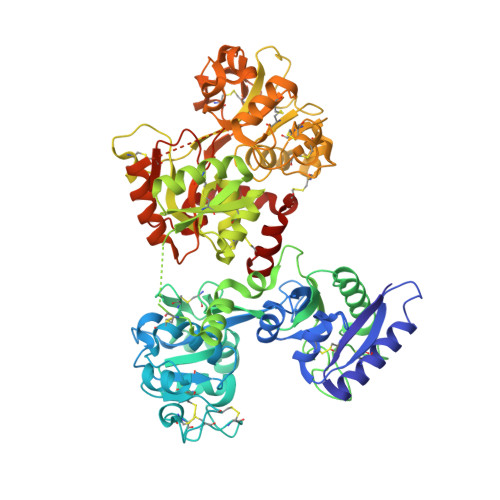
Structure of the trypanosome transferrin receptor reveals mechanisms of ligand recognition and immune evasion.
Trevor, C.E., Gonzalez-Munoz, A.L., Macleod, O.J.S., Woodcock, P.G., Rust, S., Vaughan, T.J., Garman, E.F., Minter, R., Carrington, M., Higgins, M.K.(2019) Nat Microbiol 4: 2074-2081
- PubMed: 31636418
- DOI: https://doi.org/10.1038/s41564-019-0589-0
- Primary Citation of Related Structures:
6SOY, 6SOZ - PubMed Abstract:
To maintain prolonged infection of mammals, African trypanosomes have evolved remarkable surface coats and a system of antigenic variation 1 . Within these coats are receptors for macromolecular nutrients such as transferrin 2,3 . These must be accessible to their ligands but must not confer susceptibility to immunoglobulin-mediated attack. Trypanosomes have a wide host range and their receptors must also bind ligands from diverse species. To understand how these requirements are achieved, in the context of transferrin uptake, we determined the structure of a Trypanosoma brucei transferrin receptor in complex with human transferrin, showing how this heterodimeric receptor presents a large asymmetric ligand-binding platform. The trypanosome genome contains a family of around 14 transferrin receptors 4 , which has been proposed to allow binding to transferrin from different mammalian hosts 5,6 . However, we find that a single receptor can bind transferrin from a broad range of mammals, indicating that receptor variation is unlikely to be necessary for promiscuity of host infection. In contrast, polymorphic sites and N-linked glycans are preferentially found in exposed positions on the receptor surface, not contacting transferrin, suggesting that transferrin receptor diversification is driven by a need for antigenic variation in the receptor to prolong survival in a host.
- Department of Biochemistry, University of Cambridge, Cambridge, UK.
Organizational Affiliation: